

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
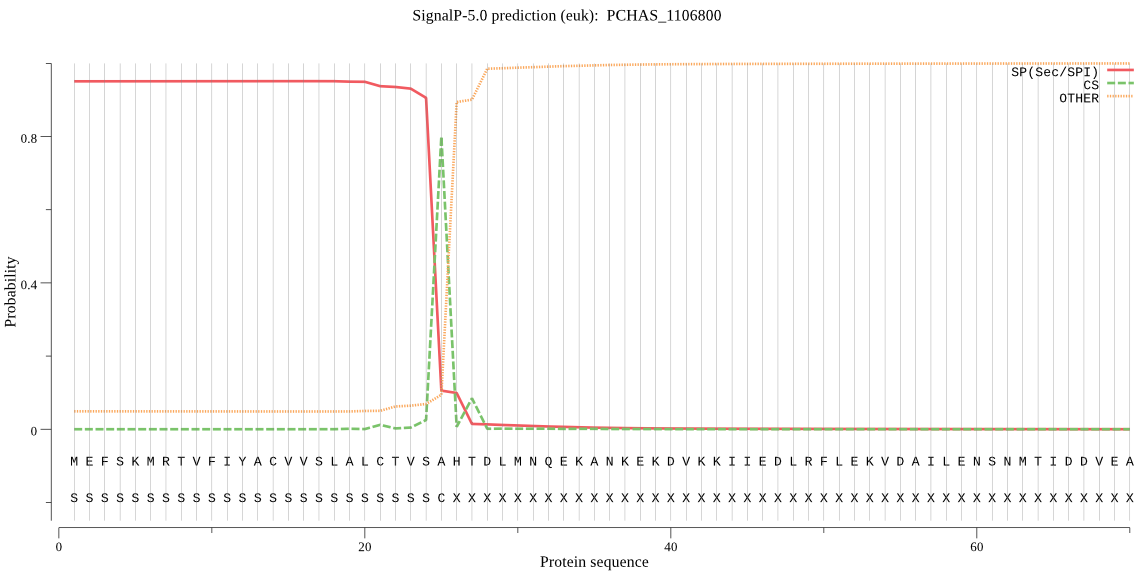
| PCHAS_1106800 | SP | 0.055646 | 0.943904 | 0.000450 | CS pos: 25-26. VSA-HT. Pr: 0.7821 |
MEFSKMRTVFIYACVVSLALCTVSAHTDLMNQEKANKEKDVKKIIEDLRFLEKVDAILEN SNMTIDDVEADDDAYNPDEDAPKEELNKIEMEKKKAEEEEKHSKKNREEKKNKSLRLIVS ENHTTSPSFFEESIIQEDFMSFIQSKGDIVNLKNLKSMIIELNSDMTDKELEDYITLLKK KGAHVESDELVGADSIYVDIIKEAVKRGDTSINFKKMHSNMLEVENKTYDKLNSKLEKPK NSKKQSVFNDEYRNLQWGLDLARLDEAHELMKDNLVEDTKVCVVDSGIDYTHPDIRDSIY VNLKELNGKSGVDDDNNGVVDDIYGANYVSNNGDPWDDHNHGTHVAGIISAIGNNSIGVV GVNPKSKLVICKALDDKKLGRLGNIFKCIDYCISKEVSVINGSFSFDEYSSVFSSTIEYL AKVGILFVVSASNCLHSLNSIPDIKKCDLSVNAKYPSVLSPRHDNLLVIANLKKKRNDEY AVSINSFYSDIYCHVSAPGSNIYSTATHRSYMELSGTSMASPHVAGIASIILSINPELTY KQVIDILKNSVVKLDSHKKKVIWGGYIDILKAVKNAVSSKNSYIRSQGISIWKKKKRN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_1106800 | 103 S | EEKHSKKNR | 0.996 | unsp | PCHAS_1106800 | 242 S | KPKNSKKQS | 0.993 | unsp |