

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_1125700 | OTHER | 0.998677 | 0.001258 | 0.000065 |
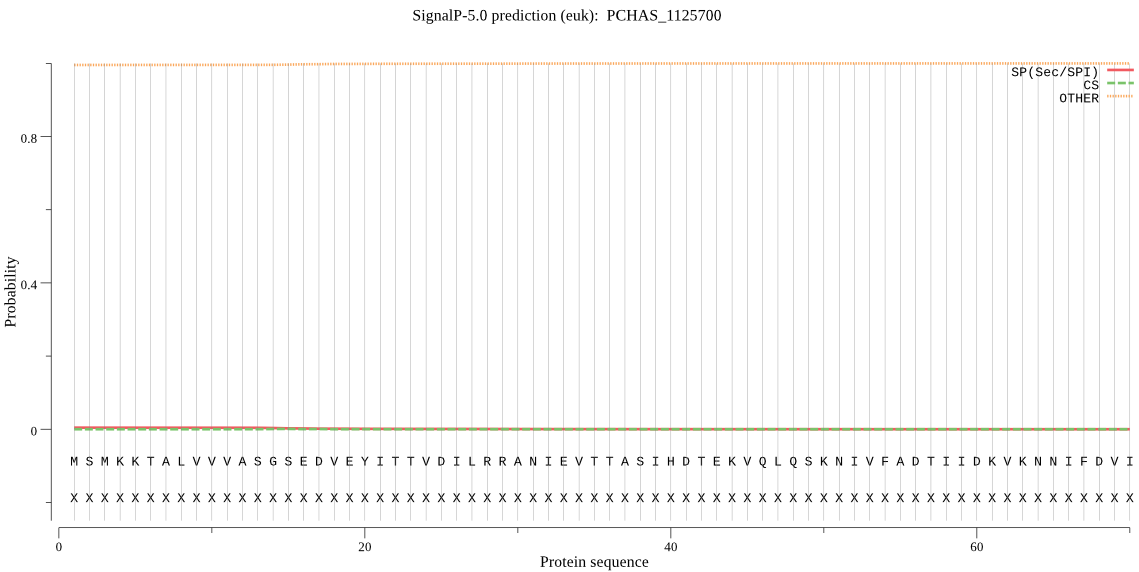
MSMKKTALVVVASGSEDVEYITTVDILRRANIEVTTASIHDTEKVQLQSKNIVFADTIID KVKNNIFDVIIIPGGMKGSNAISDCPVAIEMLREQKKNNRLYAAICAAPATVLHRHSLID DVEAVAYPSFESDFKHIGKGRVCVSKNCVTSLGPGTAGEFAFKIVELLLGRDAALRLASG FLLHPSITF
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_1125700 | 38 S | VTTASIHDT | 0.993 | unsp | PCHAS_1125700 | 117 S | LHRHSLIDD | 0.997 | unsp |