

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
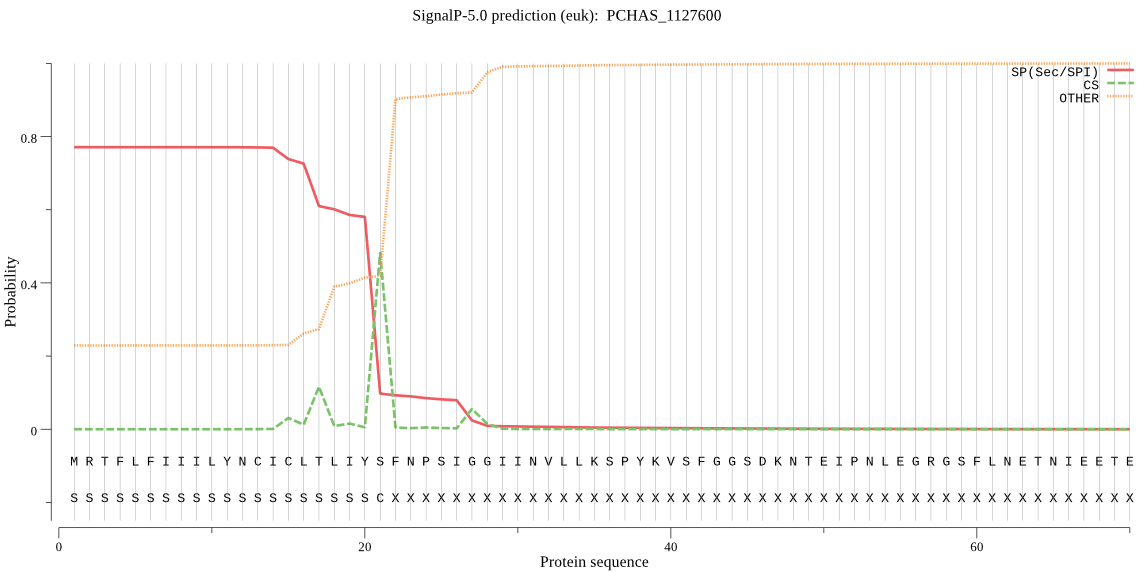
| PCHAS_1127600 | SP | 0.031187 | 0.967770 | 0.001043 | CS pos: 21-22. IYS-FN. Pr: 0.8180 |
MRTFLFIIILYNCICLTLIYSFNPSIGGIINVLLKSPYKVSFGGSDKNTEIPNLEGRGSF LNETNIEETEQIVEFESKESKNDGQDLLNNESAPELVIEREMATTKEAIVEEAAIDKETV IEETVDNDDPSDEIKRIKEVTSEEIEKIKETAAEEIEKIKETVAEEIEIAKEIIAGAVLE NKEKGEDKEPVSEGTEENKEKEEDKELISENTEENKEKEEVLKNANIENESIVEEIDKSN GKDPSKMTKEEKTNEMIQEKYGDFKKDENSYYWESTSTNAEDDAVDDVYDDAYDDIEIVG SSKENNSTISDNKNKNKNSVYLIPGLGGSTLIAEYNDAKIESCSSKVLHSKPYRIWLSLS RMSSLKSNVYCLFDTLKLDYDRENKIYVNKPGVIINVESYGYTKGVAFLDYIRNRPLRLT RYYGIIADKFLKNEYVDGKDILSAPYDWRFPLSQQKYHVLKSHIEYIYKLKNETKVNLVG HSLGGLFINYFLSQFVDDEWKKKHINIVMHISVPFAGSIKAIRALLYTNKDYTVLKLRNI LKVSISGSLMKTIAHSIGSIFDLLPYRKYYDRDQIVVIINMSELPIDEKLVQSIVTECGI YNESCYTDREGVNLKTYTLSNWHELLNDDLREKYENYIQYTDRFFSVDHGIPTYCLYSTT RKKTTEYMLFYQDAHLNQDPIIYYGIGDETISLESLEACKKLQNVKETKHFEYYKHLGIL KNSKVSDYIYNIIDQNTTN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_1127600 | 364 S | SRMSSLKSN | 0.992 | unsp | PCHAS_1127600 | 364 S | SRMSSLKSN | 0.992 | unsp | PCHAS_1127600 | 364 S | SRMSSLKSN | 0.992 | unsp | PCHAS_1127600 | 80 S | ESKESKNDG | 0.994 | unsp | PCHAS_1127600 | 142 S | KEVTSEEIE | 0.992 | unsp |