

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_1134100 | SP | 0.125084 | 0.866523 | 0.008393 | CS pos: 18-19. GNS-LH. Pr: 0.6893 |
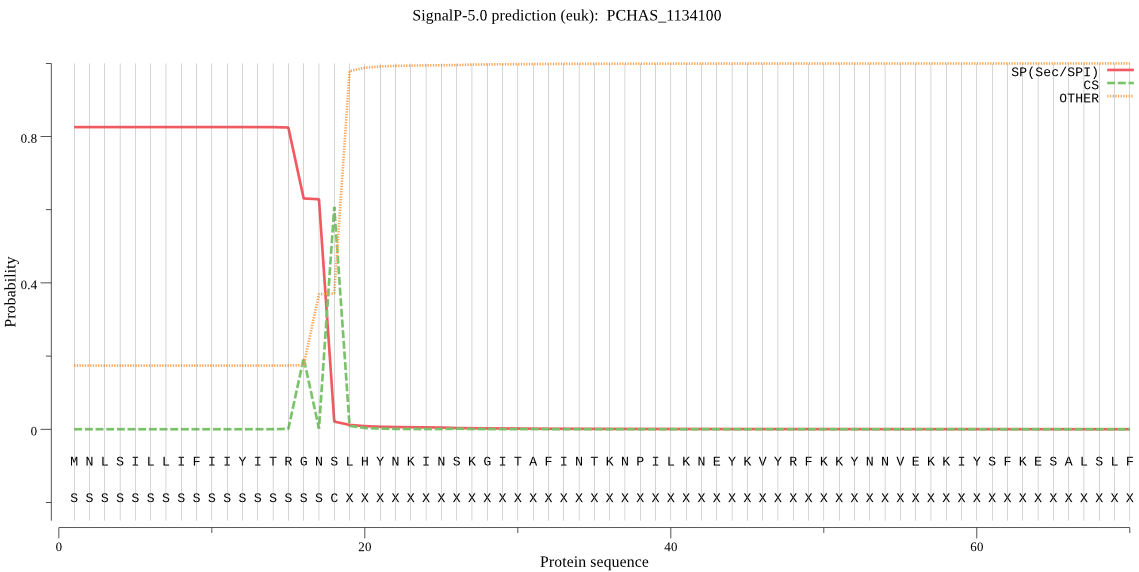
MNLSILLIFIIYITRGNSLHYNKINSKGITAFINTKNPILKNEYKVYRFKKYNNVEKKIY SFKESALSLFNTVKDKEKIANLLENISNNVKIKFPNRFIYYNYLFNKCKLDRILIVINTL LYLYLNRVDKNEEKKIFFTKGNLVQIKDEQKAEKYQCNYYDIYKNKNYKTLFSSIFIHKN ILHLYFNMSSLMSIYRMISPIYSNSQILITYLLSGFLSNLISYIYYMKPLKKDIFLKDII DQNYYSRDIPLNKPNKIICGSSSAIYSLYGMYITHMIFFYFKNNYIMNTGFLYNIFYSFL SSLLLENVSHFNHILGFISGFFMSSTLILFDNN
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PCHAS_1134100 | 61 S | KKIYSFKES | 0.994 | unsp |