

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
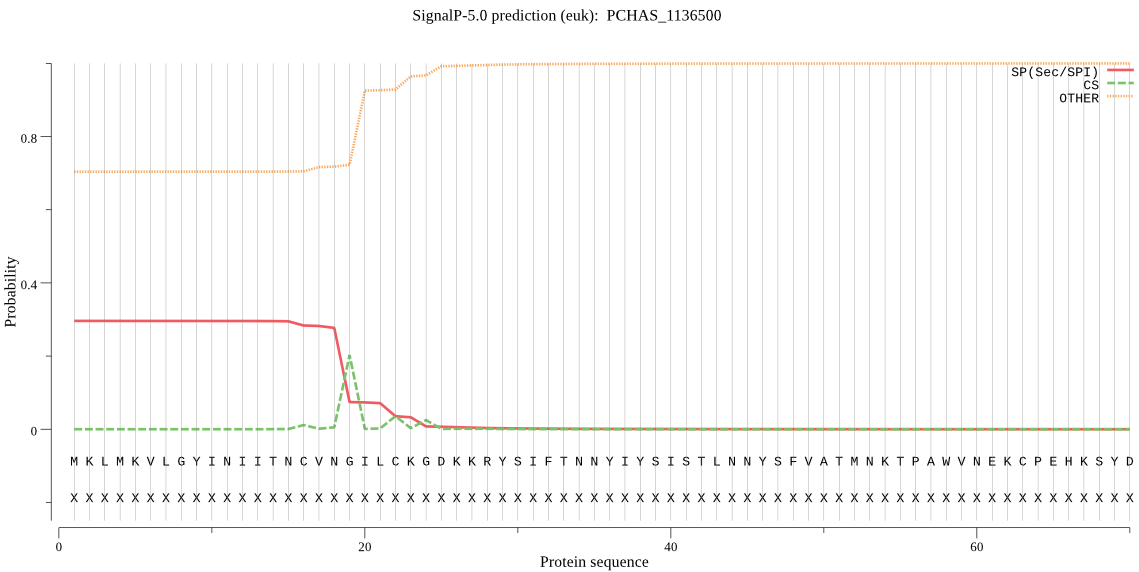
| PCHAS_1136500 | SP | 0.484340 | 0.485528 | 0.030132 | CS pos: 19-20. VNG-IL. Pr: 0.7492 |
MKLMKVLGYINIITNCVNGILCKGDKKRYSIFTNNYIYSISTLNNYSFVATMNKTPAWVN EKCPEHKSYDIVEKRYNENLNLTYTVYEHKKAKTQVIALGSNDPLDVEQAFGFYVKTLTH SDKGIPHILEHTVLSGSKNFNYKDSMGLLEKGTLNTHLNAYTFNDRTIYMAGSMNNRDFF NIMAVYMDSVFQPNVLENKFIFQTEGWTYEVEKLKEEEKNLDIPKIKDYKVSFNGIVYNE MKGAFSNPLQDLYYEVMRNMFPDNVHSNVSGGDPKEIPNLSYEEFKEFYYKNYNPKKIKV FFFSKNNPTELLNFVDKYLCQLDFTKYRDDAVEHVNYQEYRKGPFYIKKKFADHSEEKEN LASVSWLLNPKKHKNSDADLSLESPTDYFAWLIINNLLTHTSESVLYKALIESGLGNSIV DRGLNDSLVQYVFSIGLKGIKEKNEKNISLDKVHYEVEKIVLEALKKVVKEGFNKSAVEA AINNIEFVLKEANLKISKSIDFVFEMASRLNYGKDPLLIFEFEKHLNVVKDKIKNEPKYL EKYVEKHLLNNDHRVVILLEGDENYGAEQDKLEKDMLKKRIESFTEKEKENIITDFENLT KYKNTEESPEHLDKFPIISISDLNEKTLEIPVNPFFTNLNNENNMQNYNKTKDNQKLLKE NMDRFINKYILNKDGNDSKNADVPMLIYEIPTSGILYLQFIFSLDNLTLEELSYLNLFKS LILENKTNKRSSEDFVILREKNIGNMMANVALLSTSDRLNVTDKYNAKGFFNFEMHMLSH KCNDALEIALEALKDSDFSNKKKVIEILKRKINGMKTTFASKGHSILIKYVKSRINSKYF AYDLIHGYDNYLKLQEQLKLAETDYESLEAILNRIRKKIFKRNNLIMNVTVDPGTIDQLF AKSKNSFNNLLSYFEENESECSKDDSCNNVVGWNKEIQEKKLLEGGEKKKELLVVPTFVN SVSMSGVMFNKGEYLDPSFTVIVAALKNSYLWETVRGLNGAYGVFADIEYDGTVVFLSAR DPNLEKTLQTFREAAQGLRKMADVMTKNDLLRYIINAIGTIDRPRRGVELSKLSFSRIIS NETEQDRIEFRNRVMNTKKEDFYKFADLLEKKVKEFEKNIVVITNKEKANEYINNVDKDF KQILIE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_1136500 | 281 S | IPNLSYEEF | 0.994 | unsp | PCHAS_1136500 | 281 S | IPNLSYEEF | 0.994 | unsp | PCHAS_1136500 | 281 S | IPNLSYEEF | 0.994 | unsp | PCHAS_1136500 | 376 S | KHKNSDADL | 0.992 | unsp | PCHAS_1136500 | 384 S | LSLESPTDY | 0.996 | unsp | PCHAS_1136500 | 583 S | KRIESFTEK | 0.995 | unsp | PCHAS_1136500 | 608 S | NTEESPEHL | 0.996 | unsp | PCHAS_1136500 | 731 S | TNKRSSEDF | 0.997 | unsp | PCHAS_1136500 | 732 S | NKRSSEDFV | 0.994 | unsp | PCHAS_1136500 | 121 S | TLTHSDKGI | 0.992 | unsp | PCHAS_1136500 | 270 S | HSNVSGGDP | 0.993 | unsp |