

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_1205800 | OTHER | 0.999899 | 0.000020 | 0.000082 |
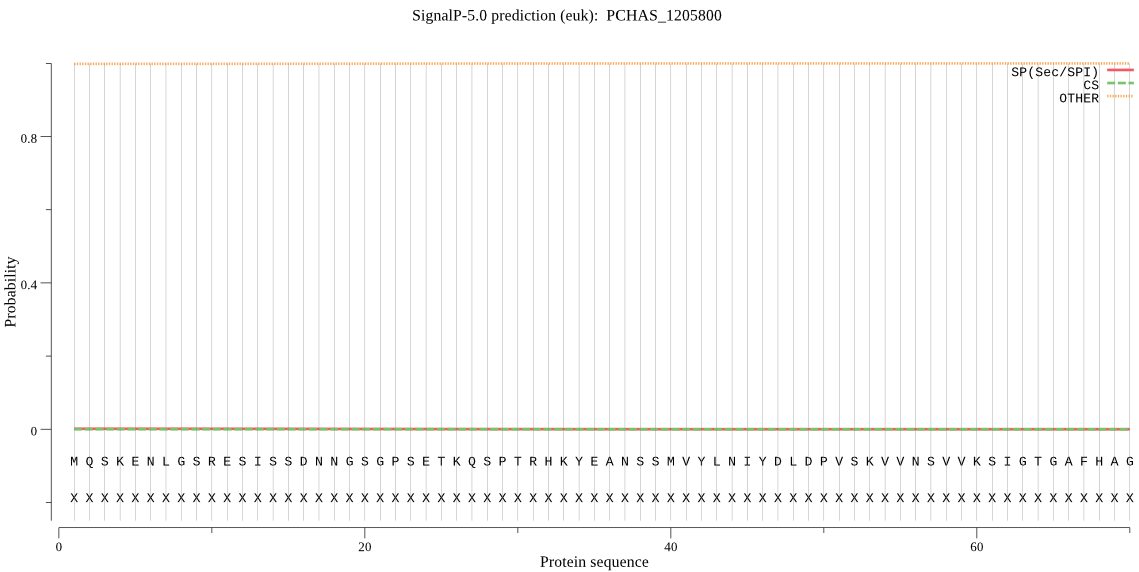
MQSKENLGSRESISSDNNGSGPSETKQSPTRHKYEANSSMVYLNIYDLDPVSKVVNSVVK SIGTGAFHAGVEVYGFEYSFGYIPGGQTGVMKTLPRHHPYHVYRESIPMGQTPLTRSEVD LLVDVMRLQWIGNTYDILSRQDIMKKYIKKSFQYHKNCLNFADYFCNLLDVGSIPEWVMS LQKNLNWMKSNISMASSKLKELNKASGLPNVLNYVKKKYNKSDESPKKCKVVLK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_1205800 | 28 S | ETKQSPTRH | 0.995 | unsp | PCHAS_1205800 | 225 S | KSDESPKKC | 0.995 | unsp |