

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
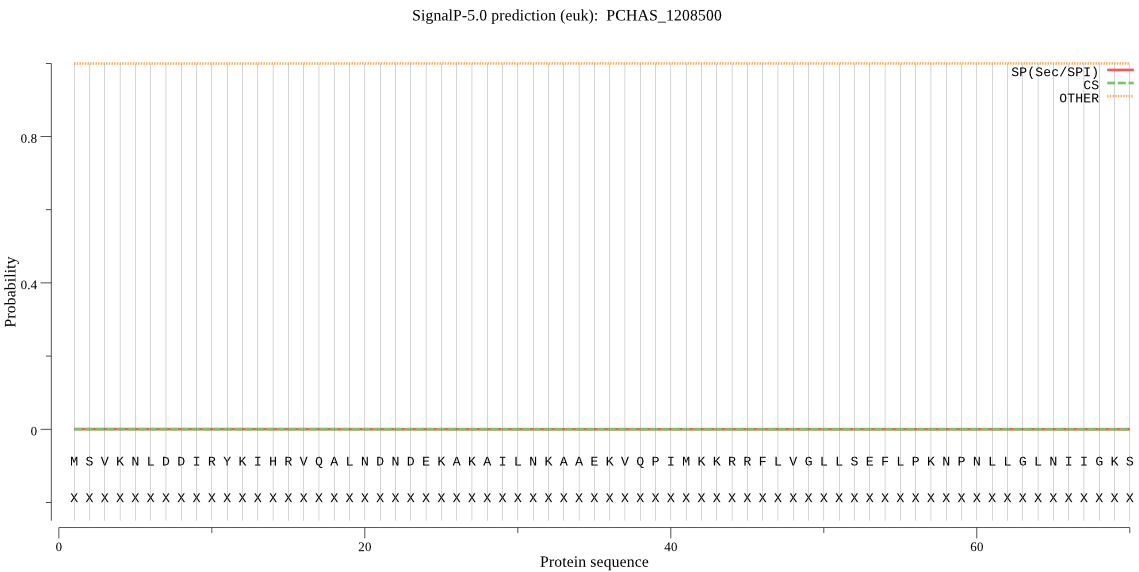
| PCHAS_1208500 | OTHER | 0.999906 | 0.000047 | 0.000048 |
MSVKNLDDIRYKIHRVQALNDNDEKAKAILNKAAEKVQPIMKKRRFLVGLLSEFLPKNPN LLGLNIIGKSEIKIRLRKKPGGEIFHFNDIMGTLLHELVHLVHRRHDKKFYALLDKLVFE YNELYIYNNINNSYGDNKNKSLANNIDENSDSYKNNASSKNNLIILIDDDFKINNHKKSS LCYSQPKHMAAQAAERRLINNFINNDGEIINISLESCLTEKQRENLIKRKKEYDDKTCLV NNDVIIIDSMSTIPKNNKRKKLTELENSNYKKNNGDNNFCNNNDCLEIISSNQPNTKNKK TLKSCSKNLDITTPECISIISDNITHIKSQKNIVEYILDDNTNTDINSTSRQ
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PCHAS_1208500 | 152 S | ENSDSYKNN | 0.994 | unsp |