

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_1220900 | OTHER | 0.998811 | 0.000938 | 0.000252 |
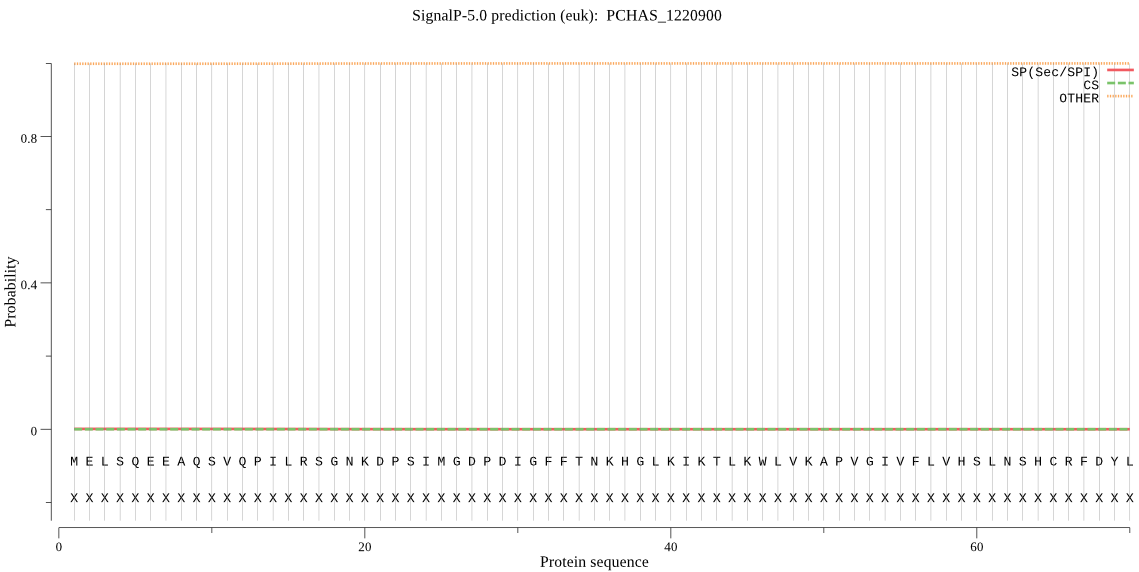
MELSQEEAQSVQPILRSGNKDPSIMGDPDIGFFTNKHGLKIKTLKWLVKAPVGIVFLVHS LNSHCRFDYLKHNVTIPNNDKAILNDGENFYVHKDSWIEELNKNGYSVYGLDLQGHGESE CYKNKKTYINEFDDFADDILQYMNGVHESIVNEAAKDNQENDNNAIGQKKRCTKCDLILK DESDICCSNEDKIIANGLYLRHPEKNQYKKPDYNIDPYKTPIPMYLVGLSMGGNIVLRIL ELLSQRRYKYYNRLNIKGACSLSGMLSAKDVRKKPSWKYFYIPLMKLSSVFFPKSRFAPN TTYEAFPFLKDLYGYDKIFYDKPITNKFVCKLLEAADNLNKDADKVVKDVPILIVHSAND QKCSHDSAQEFYNNLKTKKKEFHTLDDMEHMITMEPGNEQVIKKLVDWMAGTQKETPKKK KVKKNLRSADDKSAASSEASPTQSTSALTDLPKDKATQDQVSQEQPAEEQATEEQPVEEQ PIEEQPVEEQPIEEQPVEEQATEDQSAEEQDTQEQPTEEHDTQEQPAEEHDTQEQPTEEH DTQEHPAEEHDTQEQPTEEHDTQEHPAEEQDTQEQPAEEQDAQDQSAEEQDIQEQPAEEQ DIQEQPAEEQAIQDQSVEEQAAEEQAAEEQAAQDQAAQEQPDEEQALENEAA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_1220900 | 412 T | WMAGTQKET | 0.994 | unsp | PCHAS_1220900 | 412 T | WMAGTQKET | 0.994 | unsp | PCHAS_1220900 | 412 T | WMAGTQKET | 0.994 | unsp | PCHAS_1220900 | 436 S | KSAASSEAS | 0.996 | unsp | PCHAS_1220900 | 462 S | QDQVSQEQP | 0.995 | unsp | PCHAS_1220900 | 506 S | TEDQSAEEQ | 0.997 | unsp | PCHAS_1220900 | 586 S | AQDQSAEEQ | 0.995 | unsp | PCHAS_1220900 | 616 S | IQDQSVEEQ | 0.994 | unsp | PCHAS_1220900 | 96 S | VHKDSWIEE | 0.992 | unsp | PCHAS_1220900 | 267 S | SGMLSAKDV | 0.99 | unsp |