

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
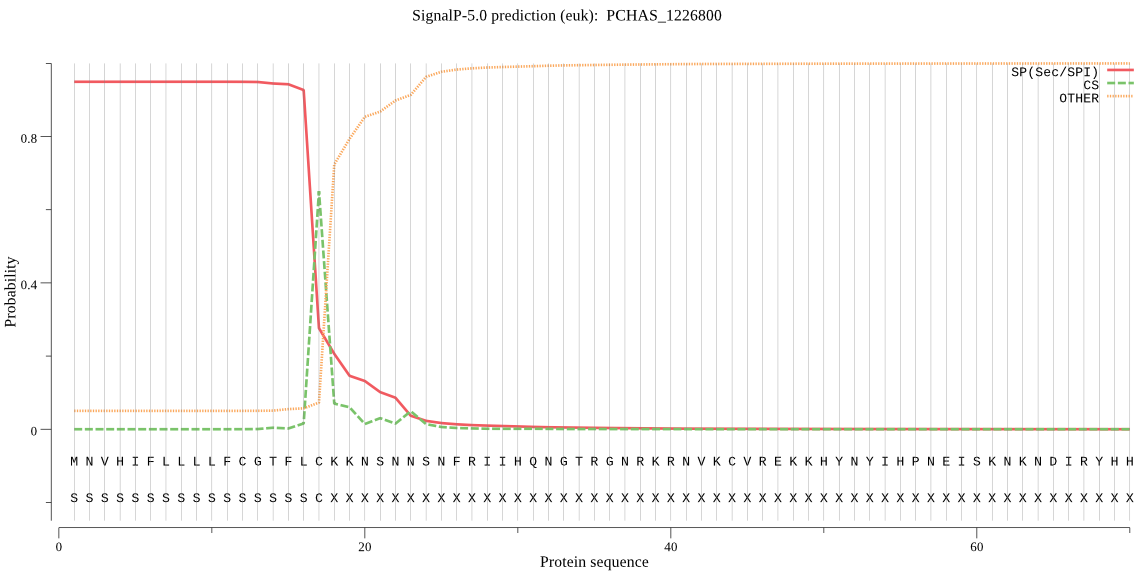
| PCHAS_1226800 | SP | 0.019516 | 0.979594 | 0.000891 | CS pos: 17-18. FLC-KK. Pr: 0.7512 |
MNVHIFLLLLFCGTFLCKKNSNNSNFRIIHQNGTRGNRKRNVKCVREKKHYNYIHPNEIS KNKNDIRYHHYNFLITTCPQKKKLLAKKEKSKRQYNLNSSGEGFSNTVHTNVKKNTNDYS EEQLPTYHKYIENFKTRKIIHPSIRITDHDKKFMKCKESYNKLYSHLTETELFENFSYVG RQRKGILSPKYYLPKYIERPNYHKTGTPIYVNYENNSKSKECISMRNSNNETENGHKTYS TTNKYEYDNVKTDYDIEMISRNCKFARELMDDISYIICEGITTNDIDIYILNKCVNNGFY PSPLNYHLFPKSSCISINEILCHGIPDNNVLYENDVVKVDISVYKDGYHADMCESFIVQK ISKEEKKKRKKNYDYIYLNDKLRTKHTKYILKYNFDLTTNKIVKKGKYPSVRKVRYNPPN SKEHENEYIEEYDDNTNSVLGESPYAHSYYTHNGGPNNVSENYEDELENFHRRYDEDFIF NPKKGEVYNNLQQYIYQKGMEKDKNRKETNRKFEFFDTSNQGIANMKKFMFEKNRDLIKT AYDCTMEAISICKPGVPFKNIAKVMDDYLKKKNNSYQYYSIVPNLCGHNIGKNFHEEPFI IHTLNDDDRKMCENLVFTIEPIITERSCDFITWPDNWTLSNSRYYYSAQFEHTILITKNG AKILTQKTDTSPKYIWEDEGN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_1226800 | 228 S | SMRNSNNET | 0.996 | unsp | PCHAS_1226800 | 362 S | VQKISKEEK | 0.996 | unsp |