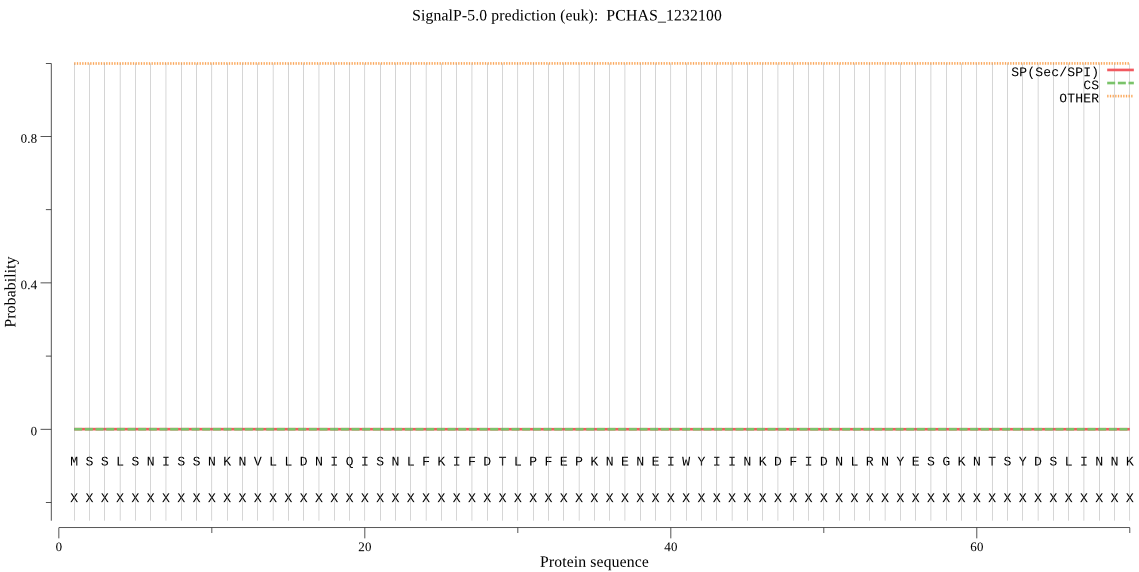
Fasta :-
MSSLSNISSNKNVLLDNIQISNLFKIFDTLPFEPKNENEIWYIINKDFIDNLRNYESGKN
TSYDSLINNKIFIEDCDSDVNIRTRLLKEYENGNGISFVLYPEKAIEILEKHFQFDVKIP
RSVYQHKTKRKDKIIFKVDIQPLKVVVYSKNTNEHSNPPQMKIVILRSDTIENVLKEIIN
DFDPQNFKKHLFYAEDLGENKEKNWNCLYISNSNDYPLDKKVYEYDIDEMSCLLITNERV
DVKCLTYDNEYSEYNMTSNNLSGSVDNDTPNNPKSINYIFENGGDRGLINLGNTCFLNSS
LQCLSKILKFTNYFLSGTFWDHINYCNPVGQHGRLAKAYYETLLEMWKINKKGKPYAPKA
LKQAISEKRDEFYGYQQHDSQELLAFLLDGLHEDLNLIQKKPYYEEKLQGGLDRPDIEVA
EASWKRHKEINNSIIVDLFQGQYRSRLQCPQCNKLSITFDPFMYLSIPLPPKKNHRIWFH
VMLSREVQISLRFSSIFSGSQEVLKLKQYFINIIKSLKNKRNSILLHTKNIIKNNKYSHE
NANAFNFHTKFMDETNDNYETSNEGSPGNGMNNKNNIDGSNYMRALKNKSKIKNTLNEED
INNIYDDICHMCNINTIDDVEVGNLFFLYTRFKNLACNNFFQILNNNDLIAEPHARTGKG
ISHIFVFMVPQNWPHLIDNNKNGNNYDESGKIRTPDSLISSTGSVGKVRNNNLKDEDMYV
KNESPSSIDKKYNQNKSHSNKLGKKRKGSLTSSSNPLRKGEKSHLDNEIGKLDDLENSND
TTTEDTKNGIPNNSSNIDEGNSMEDPKVADLEKEKFTNSSNNTSLSNKPLYLLIVPLCKD
EQLLCKMKNNDLPLLVNTTCNMTAKELYDKVKSAYKKNSKSDSNKMNQNRSRSNSVNSPS
YGNGYGEKRLKNGDTEIGYYNENDNNNKLYHKNKNISNFDSSKKDNSPDIGLNYSINSNS
TNIEENSNFYENKNTNDYCESYLDKIQLYIPSYNLKENWDAKDNLGFALKYDETYISDYI
QITEENKLFIIHLDSNSIKEELSVDIKTLTLMDLEEKYSVDTCLKLFSEEEHLDEDNTWY
CSNCKLHVQAYKKLDLFRMPIVLILHLKRFNNSNRWLRTKIDSYVYYPHKENEYLDMAPY
ILEDGLKHMKSFDPSYLPLYELIGVNCHTGGLGGGHYFAYAKLNGQWYNFNDTYVTTIDE
SQINTKNAYLLFYQLHTYKNEKFTDIAQHRNIAEEEAIRMANASYYN