

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
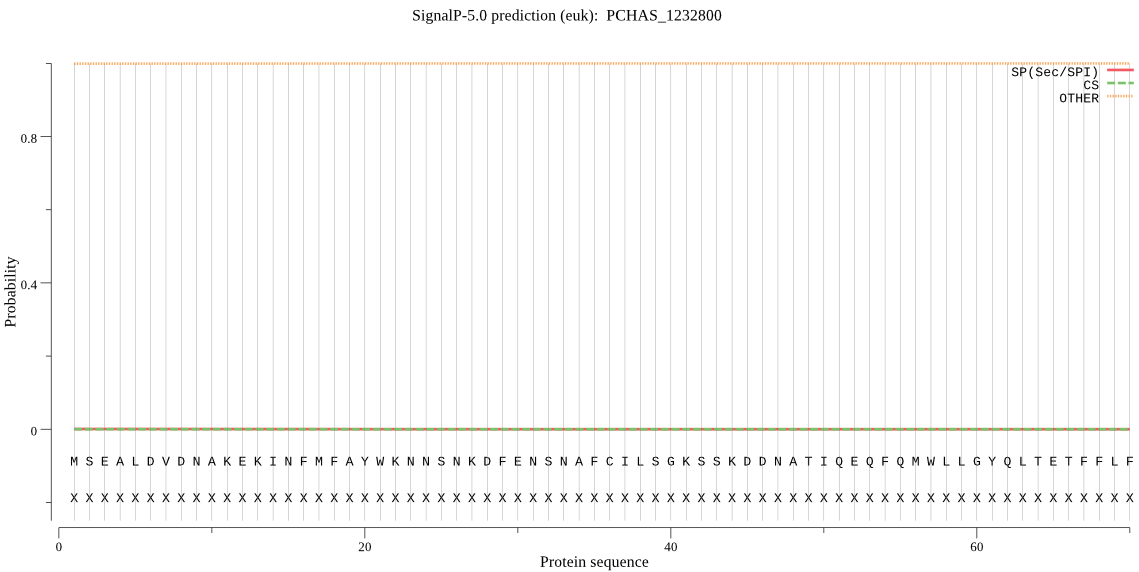
| PCHAS_1232800 | OTHER | 0.999992 | 0.000006 | 0.000002 |
MSEALDVDNAKEKINFMFAYWKNNSNKDFENSNAFCILSGKSSKDDNATIQEQFQMWLLG YQLTETFFLFCKKNEKLIILTSDKKKKFLQPLLDKTDNITILERNNNDNSENFEKIKNEI NMHDSKELLILKDKDSTGSFFEACYNFIKSLNKNEIDVNNNIKNLLNLRSKSDVKLQKSA SDIASIIMKSVLITTIENSLDSEEYESHNKIKEKVLKFNENKKCVVKIKDKLKADIDDID VIYSSVQSGNKFVLNFKNTNDNNYLSQNDGTIVIGLGVKYKELCANINRTLLLNAKEYHK ELYNFTFSLQKYIINDCLKYNTSFSDVYKKAIQYIKDNKQNYQTIGNINLENYFIKCLGH VIGFEFMEKEFLITANNSNATIEKNTSYNISVGFENVQLPDSKNVFSTWISDTVFVNDKE EVTILTDAIGKEINTISYELEESESENEEEDSEDNKKKSGKESKNVKREASDYDESDDDD DDSNNKSAKVKKEKNKNGDSENKKKIGISASILNNASSVIVSDRLRRRNKNSLAHNNEQE IEELNKRQNELKNKKIEEIKNRFSEGTNEYKDLNKKNIKKLEDIKSYNDADLIPRDLRSN IIHVDNKHESILLPVNGAHIPFHVSTIKNLSSNYEDNNDIFVLRINFQVPGNQGSQKGEF NAFPKLNEKEMYIKELIFKSNDEKHLQFVVKQVKELIKQVKQKEVEADVNDSKTSNEKLA LNKSGRRIVLRDLMTRPNIFTGRKILGTLELHTNGLRYSANSRGTTEFIDILFDDIKHAF YQPCDGQLIILIHFHLKRYIMVGKKKTLDVQFYCEVGTQIDDLDRAKARNVYDPDEMHDE MKEREQKNKLNLIFKNFVQQMQDISKIEFEIPYPELTFSGVPNKSNVEIFVTANTINHLI EWPPFILSVEDIEIASLERVHHGLRNFDMIFVFKDYTKPVKRIDVIPIEYIDTIKKWLTT IDIVYYEGKNNLQWGNILKTILADIESFVNSKGFDGFLGEDDDEEEQSAEDEDEDDEYEV DESELSAEDDSEYDDSEEESLATESDGDEEVEEDSDDEGLSWDELEERAKKDDKKRFAYQ SDDGDDSEGYNKRKKKRN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_1232800 | 43 S | SGKSSKDDN | 0.992 | unsp | PCHAS_1232800 | 43 S | SGKSSKDDN | 0.992 | unsp | PCHAS_1232800 | 43 S | SGKSSKDDN | 0.992 | unsp | PCHAS_1232800 | 443 S | ELEESESEN | 0.992 | unsp | PCHAS_1232800 | 459 S | NKKKSGKES | 0.998 | unsp | PCHAS_1232800 | 471 S | KREASDYDE | 0.997 | unsp | PCHAS_1232800 | 476 S | DYDESDDDD | 0.991 | unsp | PCHAS_1232800 | 564 S | KNRFSEGTN | 0.993 | unsp | PCHAS_1232800 | 762 S | YSANSRGTT | 0.992 | unsp | PCHAS_1232800 | 908 S | PFILSVEDI | 0.993 | unsp | PCHAS_1232800 | 1008 S | EEEQSAEDE | 0.997 | unsp | PCHAS_1232800 | 1026 S | ESELSAEDD | 0.997 | unsp | PCHAS_1232800 | 1031 S | AEDDSEYDD | 0.997 | unsp | PCHAS_1232800 | 1036 S | EYDDSEEES | 0.995 | unsp | PCHAS_1232800 | 1045 S | LATESDGDE | 0.993 | unsp | PCHAS_1232800 | 1061 S | DEGLSWDEL | 0.995 | unsp | PCHAS_1232800 | 25 S | WKNNSNKDF | 0.992 | unsp | PCHAS_1232800 | 42 S | LSGKSSKDD | 0.997 | unsp |