

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_1304600 | OTHER | 0.999953 | 0.000021 | 0.000025 |
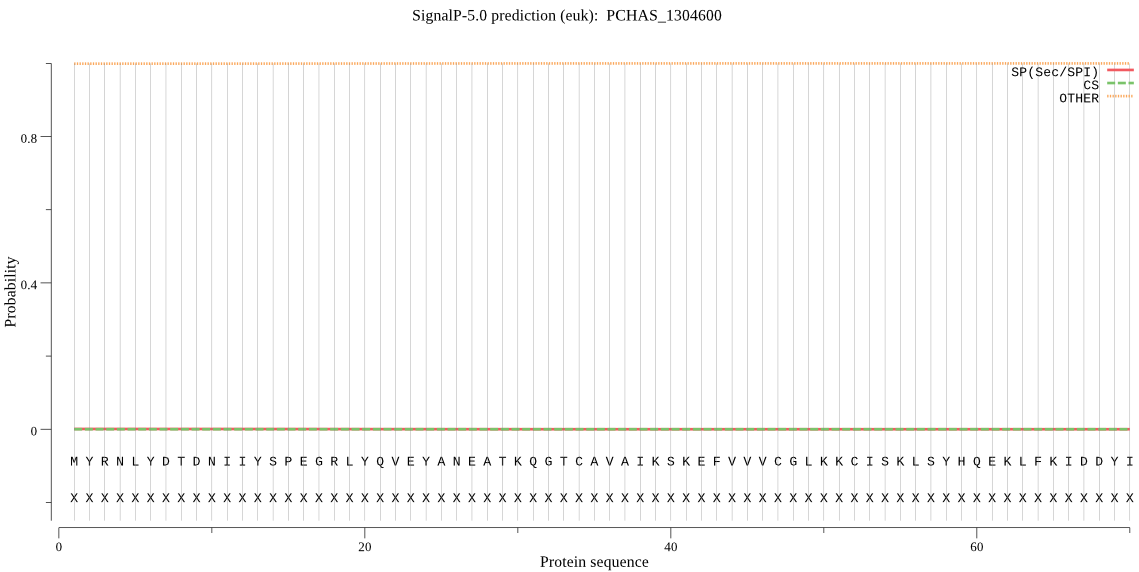
MYRNLYDTDNIIYSPEGRLYQVEYANEATKQGTCAVAIKSKEFVVVCGLKKCISKLSYHQ EKLFKIDDYIGVTMSGITSDAKVLTKYMRNECLTHKFLFDENINVEKLVKKVADKYQKNT QKSSKRAFGVGLIISGYFNEPHIFETKSNGSYFEYEALSFGARSHASKTYLEKNLHLFEE SSLDDLIIHCLKALKCSLSSETELTADNTSLAIVGKDKPWEEMPYVELIDYLTKVNSEKR TEDSQDNVENSGN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_1304600 | 237 S | TKVNSEKRT | 0.996 | unsp | PCHAS_1304600 | 237 S | TKVNSEKRT | 0.996 | unsp | PCHAS_1304600 | 237 S | TKVNSEKRT | 0.996 | unsp | PCHAS_1304600 | 123 S | NTQKSSKRA | 0.997 | unsp | PCHAS_1304600 | 151 S | KSNGSYFEY | 0.994 | unsp |