

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_1308800 | OTHER | 0.999940 | 0.000008 | 0.000053 |
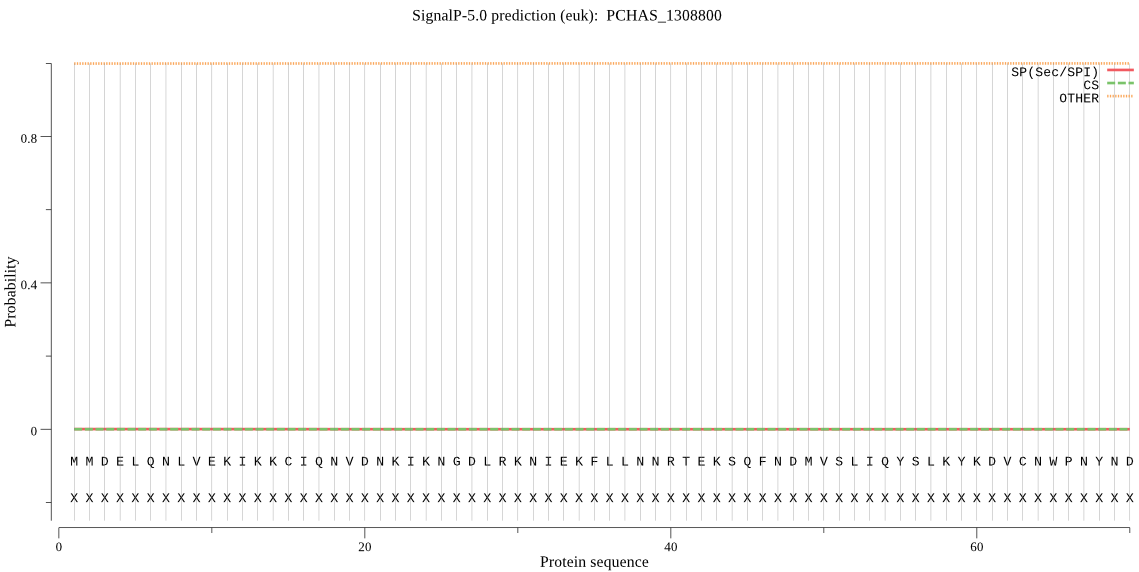
MMDELQNLVEKIKKCIQNVDNKIKNGDLRKNIEKFLLNNRTEKSQFNDMVSLIQYSLKYK DVCNWPNYNDLFIINYNINNNFKSIIQKNENFLRQYKNHTSNEYEENVTTESSIKNDMKT TNPINTDKNISLAMDNEMNNKKNVPSFTEHNKVSENNQDNIAGKGNNSMDQNLVDKIKLN SKGANTNPKLNNINIQYNDDITLFNKLKLQTFMYFVRNNYKVKLLIDSLSAMNHPINIIY INCPNNKYKKKNIFQKISDLFLPDYKVNDEFVNSKYNYQKNNCSCSELPLNPLSNNNYAY TQKKNVSNYTGGYNPISNTVWLCANNITNLYKLKYILTHELIHAFDFARANIDMYNCHHI ACSEIRAYNMSNQCGYFNSKHFLPNRDVFNHFKAPSINDTAKNKCVYNNTYTSLYQYKPC ANNTHKYINDVFEKCIHDYWPFMCAPEQDSKYKPSKT