

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_1319300 | SP | 0.012546 | 0.987345 | 0.000110 | CS pos: 18-19. ING-IQ. Pr: 0.8195 |
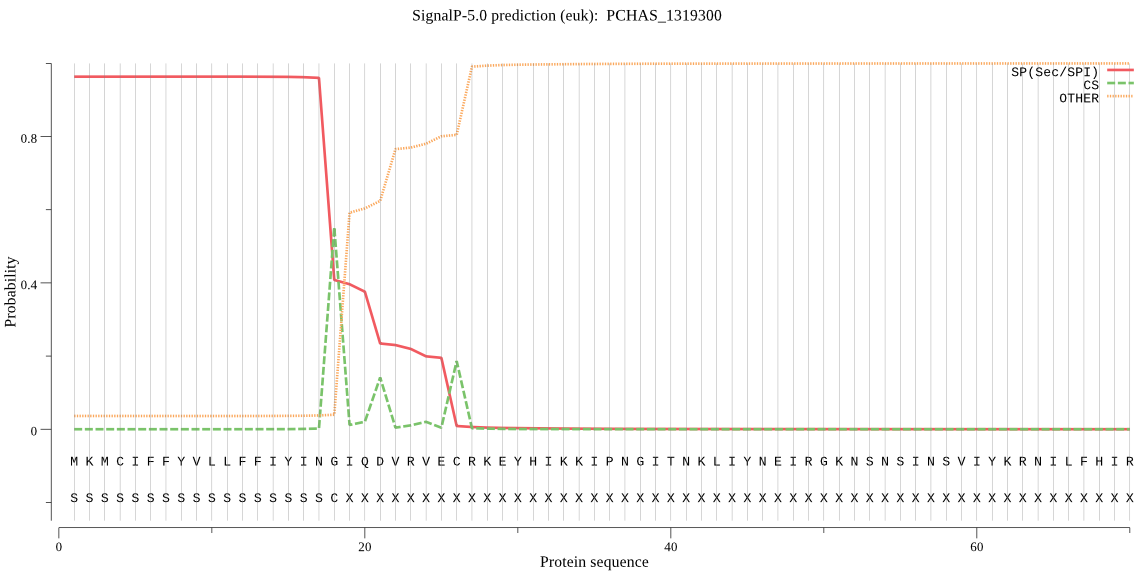
MKMCIFFYVLLFFIYINGIQDVRVECRKEYHIKKIPNGITNKLIYNEIRGKNSNSINSVI YKRNILFHIRIPKKTLQGRNLFSNDIRNKKINEFLQKKKNSCYLHINSDVNEIKKKRDKL LKDVLLNAKQLQEYINNLKSTLLHKYKDTKIITKFFLSTSFLVMVLNLFGLRPEDIALHD KRIIRAFEFYRIITSALFYGDISLYVLTNIYMLYLQSQELEKSVGSSEALAFYLSQITIL STICSFMKKPFYSTALLKSLLFTNCMLNPYNKSNLIFGINIYNMYLPYLSIVIDILHAQD FNASLSGILGVISGAIYYFSNIYLLEKCNTKFFKIPQILRNYLDSFNTDEFVF