

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
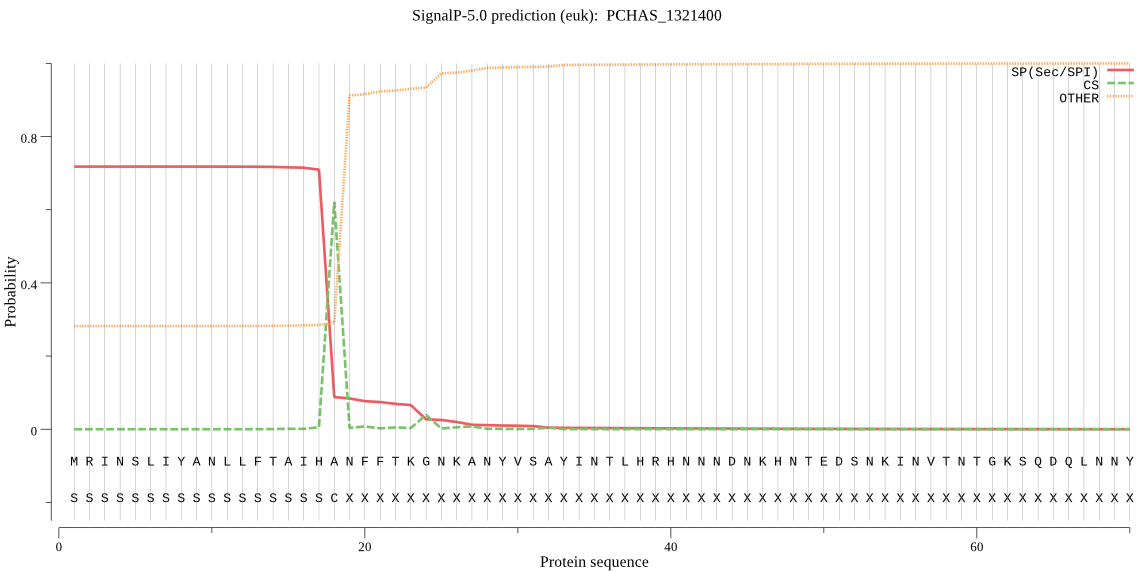
| PCHAS_1321400 | SP | 0.084262 | 0.875918 | 0.039820 | CS pos: 18-19. IHA-NF. Pr: 0.9661 |
MRINSLIYANLLFTAIHANFFTKGNKANYVSAYINTLHRHNNNDNKHNTEDSNKINVTNT GKSQDQLNNYFSRRFYTIQNKNQYLREGKRSSTDDNLTNYNMESSNEQNNQMMGGDSYKE RLQKLKKYMGDHNIDVYILINSDAHNSEIINDQDKKIYYLTNYSGADGILILTKDQQIIY VNALYELQATKELDTKFFDLKIGRITNKDEIFQTIADLEFNTIAFDGKNTSVSFYEKLKN KLKFQYPDKQIQEKFIYKDSINKIVKDKNINLYVLESPLVTVPNDNVNKKPVFIYDREFG GACAAQKIQEASDFFDENPDVDSMLLSELDEIAYLLNLRGYDYVYSPLFYSYVYLKYNRE KGRIDEIILFAKTENIKENVLAHLDRIHVKLMDYDSVVSFLTKNVSTKTANITRYNENNL LLSSLQGNSNPRYDISLSPHINLMVYMLFNKEKVLLKKSPIVDMKAVKNYVEMDSIKEAH VLDGLALLQFFHWCDEKRKTKELFKETEISLRNKIDYFRSTKKNFIFPSFSTISAIGPNS AVIHYESTEETNAKITPSIYLLDSGGQYLYGTTDVTRTTHFGEPNADEKKLYTLVLKGHL SLRKVIFASYTNSMALDFLARQPLYNHFLDYNHGTGHGVGICLNVHEGGCSISPAAGTPL KESMVLSNEPGYYWADHFGIRIENMQFVVTKKQTDDTTFLTFNDLTLYPYEKKLLDYSLL TPQEIADINEYHLTIRNTLLPRIKENPSEYDKGVEQYLMEITEPISH
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_1321400 | 117 S | MGGDSYKER | 0.995 | unsp | PCHAS_1321400 | 117 S | MGGDSYKER | 0.995 | unsp | PCHAS_1321400 | 117 S | MGGDSYKER | 0.995 | unsp | PCHAS_1321400 | 475 S | VEMDSIKEA | 0.995 | unsp | PCHAS_1321400 | 547 S | IHYESTEET | 0.99 | unsp | PCHAS_1321400 | 748 S | KENPSEYDK | 0.996 | unsp | PCHAS_1321400 | 91 S | EGKRSSTDD | 0.994 | unsp | PCHAS_1321400 | 92 S | GKRSSTDDN | 0.998 | unsp |