

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
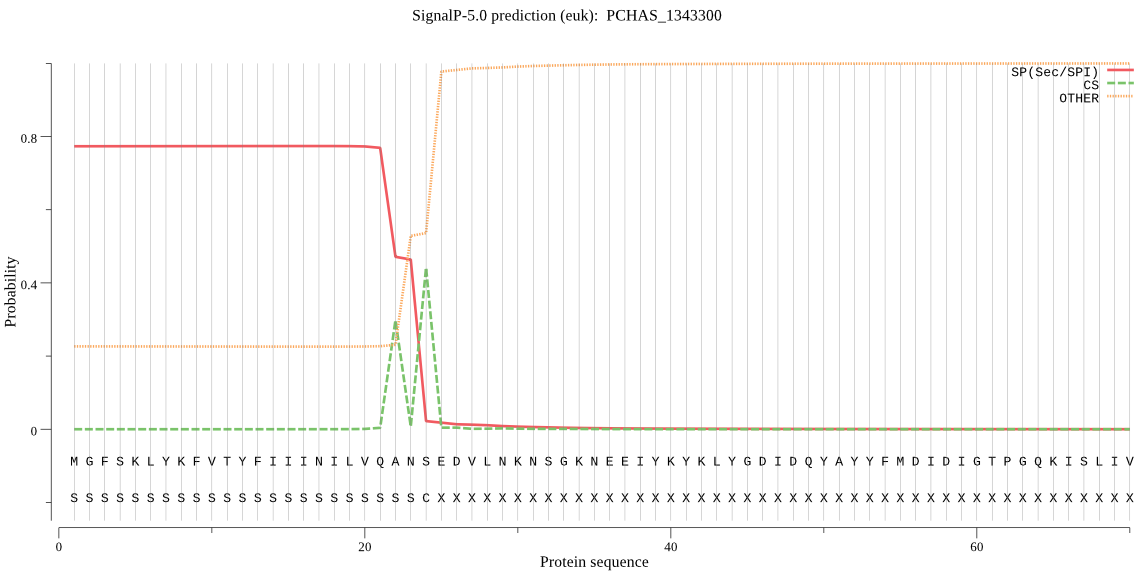
| PCHAS_1343300 | SP | 0.007454 | 0.992515 | 0.000031 | CS pos: 24-25. ANS-ED. Pr: 0.6117 |
MGFSKLYKFVTYFIIINILVQANSEDVLNKNSGKNEEIYKYKLYGDIDQYAYYFMDIDIG TPGQKISLIVDTGSSSLSFPCSECKDCGVHMENPFNLNNSSTSSVLYCNDNSNCPYHLKC VKGRCEYLQSYCEGSRINGFYFLDFVKLESYNNTNNGDIIFKKHMGCHMHEEGLFLQQHA TGVLGLSLTKPKGVPTFIDLLFKNSPKLNKVFSLCISEYGGEMIIGGYSKNYVIKDVSNN EKSDQIVHNKDEDINSVDKSMEINKNKSIVDDIVWEAITKKYYYYIKVEGFELFGTTFSH NNTSMEMLVDSGSTFTHLPDDLYNSLNFFFDLLCIHNMNNPIDIEKRLKITNETINKHIL YFDDFKSTLKNIISSENMCVKIADNVQCWRYLKHLPNIYIKLSNNTRLLWKPSSYLYKKE SFWCKGLEKQVNNKPILGLSFFKNKQIIFDLKNNKIGFVESNCPSNPINTRPRTFNEYNI KENYLYKQSYFSLYSLSIIIALTFILYIILYIKKIIYSYYNPLHGENTSSD
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PCHAS_1343300 | 268 S | NKNKSIVDD | 0.996 | unsp |