

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_1370600 | OTHER | 0.999906 | 0.000023 | 0.000071 |
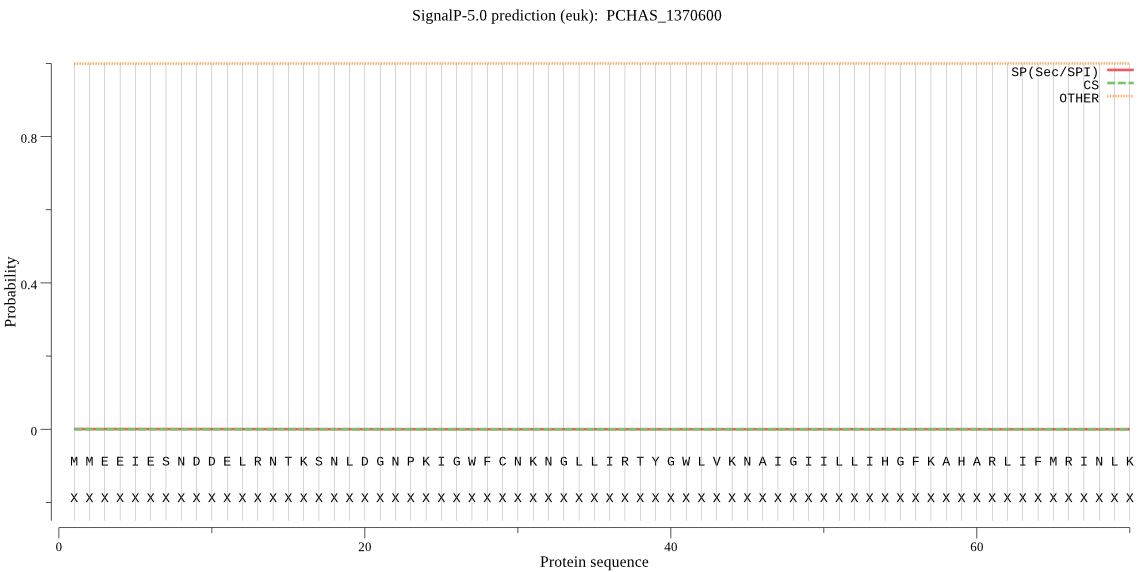
MMEEIESNDDELRNTKSNLDGNPKIGWFCNKNGLLIRTYGWLVKNAIGIILLIHGFKAHA RLIFMRINLKMPDSNEGLVVDTNNYYIYKDSWIEKFNQNSYSVYALDLQGHGESQSFENI RGNFNCFDDLVDDVMQYMNQIQDEISNDNQMNDEPHNIVTTKKKKLPMYVVGYSMGATIA LRILQLLGEEKEDNINAGDENNYKKCKSILDNSTDVNELGNDMNNYNDYGSDNSCASGNH EGRYNYLDKLNIKGCVSISGMMRMKTPLNTGNKTHKYLYLPIINFLSRVAPHILIPPITG YKKSEYIINVCKHDKFINDNGTKFKCFAELIKAMLTLDSNINYIPKDIPILFVHSKDDSI CHYKGSVSFYNKVNASGNELYPVDGMNHSTLTQPGNEEILKKVIDWISNLRTNDEDE
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PCHAS_1370600 | 355 S | LFVHSKDDS | 0.993 | unsp |