

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
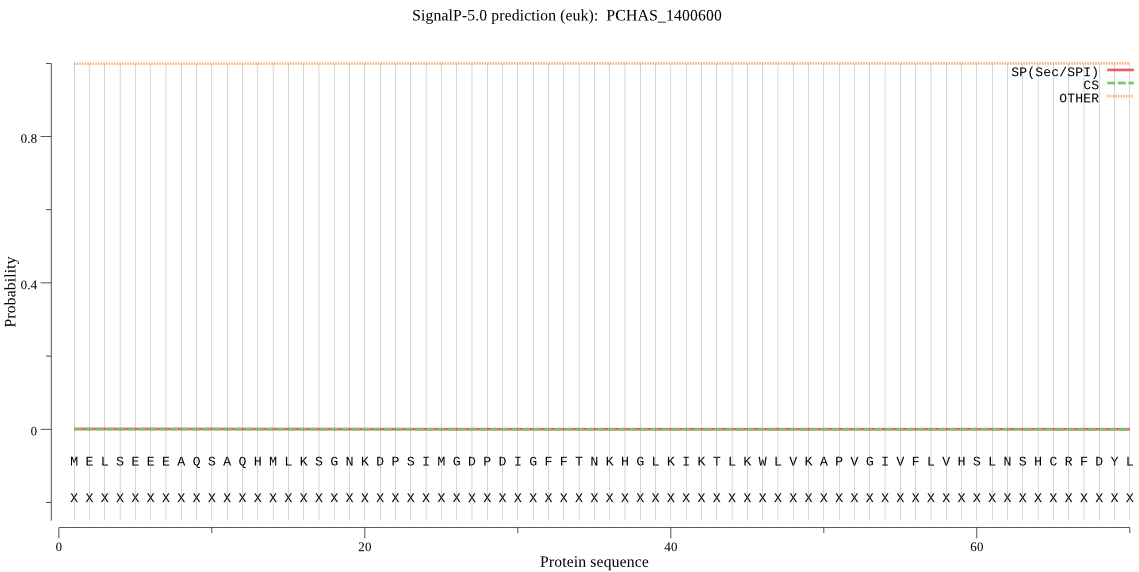
| PCHAS_1400600 | OTHER | 0.999635 | 0.000227 | 0.000139 |
MELSEEEAQSAQHMLKSGNKDPSIMGDPDIGFFTNKHGLKIKTLKWLVKAPVGIVFLVHS LNSHCRFDYLKHNVTISNNEKAVLNDGDNFYVHKDSWIEELNKNGYSVYGLDLQGHGESE CYGNEKTHINEFDDFADDILQYMNMAHESIVNEAAKDNQENDNNAIGQKKRCTKCDLILK DESDICCSNEDKIIANGLYLRHPEKNQYKKTDYNIDLYKTPIPMYLVGLSMGGNIILRIL ELLSQRRYNYYNRLNIKGVCSLSGMVSAKEVKKKFSWKCFCIPMMTLSSLLFPTSRFSSN ATSETFPYIKDLYIYDKIFDGKPITNKFAHKLLEAVDNLNKDIDKMVKDVAILLIHSAND KNCSYTAAQEFYNNLKTKKKEFHTLDDMEHMITMEPGNEQVVKKLVDWMAGTQKETPKKK KAKKNLRSADDKNILPNEASPMQGTGVLTDPPKDKTTQDQVAQEQVSQAQVSQEQVSQEQ VSQEQVSQEQVSQEQVSQEQVSQEQVSQAQVSQEQVSQEQTSQEQTSQEQVSQEQTSQEQ VSQEQTSQEQVSQEQVSQEQTSQEQASQEQTSQEQASQEQTSQEQASQEQTSQEQTSQEQ TSQEQASQEQTSQEQASQEQTSQEQASQEQPAEEQAVQDQPAEEQYTQEQPAEEQALQNE AA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_1400600 | 298 S | TSRFSSNAT | 0.99 | unsp | PCHAS_1400600 | 298 S | TSRFSSNAT | 0.99 | unsp | PCHAS_1400600 | 298 S | TSRFSSNAT | 0.99 | unsp | PCHAS_1400600 | 412 T | WMAGTQKET | 0.994 | unsp | PCHAS_1400600 | 517 S | QEQVSQEQT | 0.995 | unsp | PCHAS_1400600 | 522 S | QEQTSQEQT | 0.996 | unsp | PCHAS_1400600 | 532 S | QEQVSQEQT | 0.995 | unsp | PCHAS_1400600 | 542 S | QEQVSQEQT | 0.995 | unsp | PCHAS_1400600 | 557 S | QEQVSQEQT | 0.995 | unsp | PCHAS_1400600 | 562 S | QEQTSQEQA | 0.995 | unsp | PCHAS_1400600 | 567 S | QEQASQEQT | 0.995 | unsp | PCHAS_1400600 | 572 S | QEQTSQEQA | 0.995 | unsp | PCHAS_1400600 | 577 S | QEQASQEQT | 0.995 | unsp | PCHAS_1400600 | 582 S | QEQTSQEQA | 0.995 | unsp | PCHAS_1400600 | 587 S | QEQASQEQT | 0.995 | unsp | PCHAS_1400600 | 592 S | QEQTSQEQT | 0.996 | unsp | PCHAS_1400600 | 597 S | QEQTSQEQT | 0.996 | unsp | PCHAS_1400600 | 602 S | QEQTSQEQA | 0.995 | unsp | PCHAS_1400600 | 607 S | QEQASQEQT | 0.995 | unsp | PCHAS_1400600 | 612 S | QEQTSQEQA | 0.995 | unsp | PCHAS_1400600 | 617 S | QEQASQEQT | 0.995 | unsp | PCHAS_1400600 | 622 S | QEQTSQEQA | 0.995 | unsp | PCHAS_1400600 | 627 S | QEQASQEQP | 0.992 | unsp | PCHAS_1400600 | 96 S | VHKDSWIEE | 0.992 | unsp | PCHAS_1400600 | 276 S | KKKFSWKCF | 0.994 | unsp |