

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_1402100 | OTHER | 0.999645 | 0.000073 | 0.000282 |
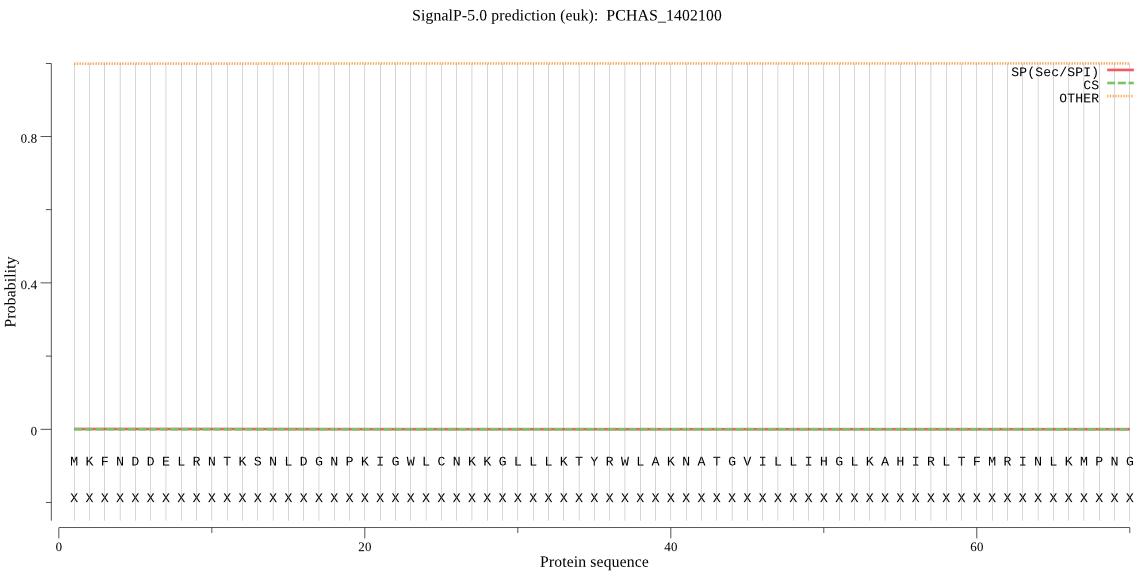
MKFNDDELRNTKSNLDGNPKIGWLCNKKGLLLKTYRWLAKNATGVILLIHGLKAHIRLTF MRINLKMPNGNEGLIVDTNNYYIYKDSWIEKFNQSGYSVYALDLQGHGESQGWKNVRGDF SSFYDLIDDVLQYMNQIQDEISNDNQTDGESYDIVSTKKKKLPMYVIGYSTGGNIALRIL QLLNKEKEDRIKAWNSNNYKNSNIILDNSTDVNDIDNGMSNYKNGDPGTSSASTSATTNA IISANDKHKGCYNSLDKFNIKGCITISGMIRIKSKRDPGNKSLKYFYLPIIKLLSLILPY KRFLPLSSYKGSAYIANVSKHDKFRNNNGITFKSMYELIKATITLDYNINYMPKDVPLLF VHSKDDRVCSYEWTALFHNKVKVDKKDLYTLDDMGHAITMKPGNEEVLKKIIDWIYDLRN GEDEIEDEI
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_1402100 | 274 S | IRIKSKRDP | 0.997 | unsp | PCHAS_1402100 | 370 S | DRVCSYEWT | 0.994 | unsp |