

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_1412200 | SP | 0.193178 | 0.802607 | 0.004215 | CS pos: 27-28. VKK-NT. Pr: 0.3127 |
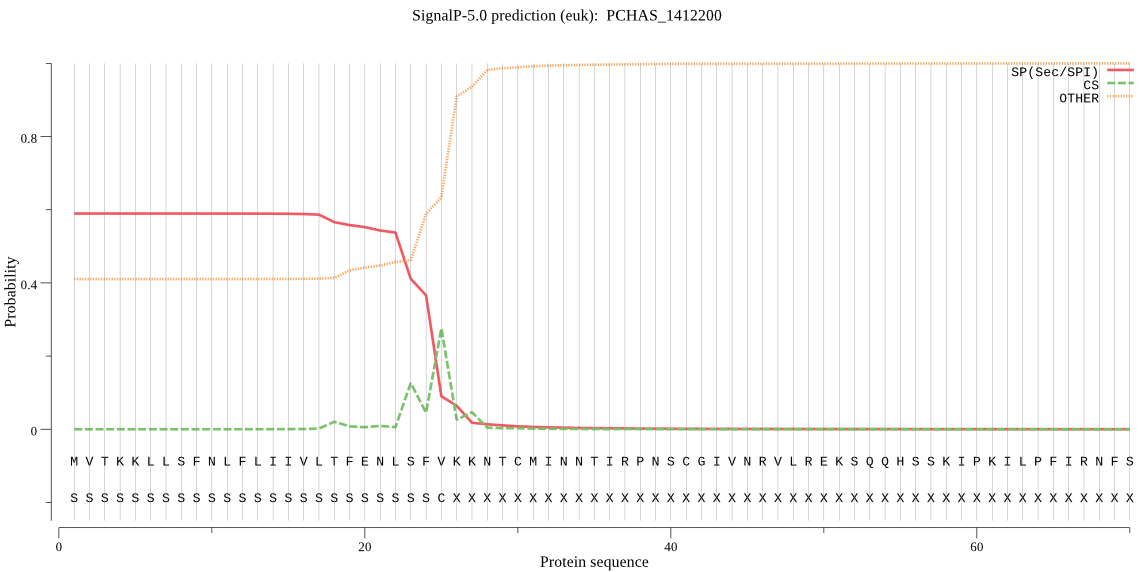
MVTKKLLSFNLFLIIVLTFENLSFVKKNTCMINNTIRPNSCGIVNRVLREKSQQHSSKIP KILPFIRNFSLKKYITGDQLQKNILNHINKLSGAHLFQGSKSQLSSITNNKFVGDATKSA LGFVSNIGTDISGNKKTNLGRMLCEGHNNNGGEVTSTENAILKNSKDPQIHYRTDYKPSG FTIDTVTLNINIFDNETTVRSSLSMCTNDNYANEDLVFDGVGLSIKEININDNKLTEGED YTYDNEFLTVFAKNVPKGNFVFVSEVIIHPETNYALTGLYKSKDIIVSQCEATGFRRITF FIDRPDMMAKYDVTLTADKTKYPVLLSNGDKLNEFDIPGGRHGARFNDPHLKPCYLFAVV AGDLKFLSDKYVTKFTKKPVELYVYSEEKYVSKLKWALECLKKAMKFDEDYFGLEYDLSR LNLVAVSDFNVGAMENKGLNIFNADSLLASKKTSIDFSFERILTVVGHEYFHNYTGNRVT LRDWFQLTLKEGLTVHRENLFSEQTTKTATFRLTHIDILRSVQFLEDSSPLSHPIRPESY ISMENFYTNTVYDKGSEVMRMYQTILGDEYYKKGISIYLKKHDGGTATCEDFNEAMNEAY QMKNGNKEENLDQYLLWFSQSGTPHVTAEYSYDANAKTFTIKLSQVTYPDDNQKEKFPLF IPVKVGLISPKDGKDVIPEVVLEFKKDKDTFVFENIEEKPIPSLFREFSAPVYIKDNLTD EERIILLKYDSDAFVRYNVCIDLYMKQIIKNYNEFLSQKTKEANGLEHSLTPVSEDFINA IKHLLEDKHSDPGFKAYIIALPRDRYIMNYIKEVDPIILADTKDYIYKQMGNRLNPILFS IFQDTESKANDMTHFKDESYVDFDQLNMRKLRNSIMVMLSKAQYPHMLKYVKDQAQSPYP SNWLASLSASAYFTGDDYYNLYDKTYNLSKNDELLLQEWLKTVSRSDRSDIYNIIKKLET EILKDSKNPNNIRAVYLPFTSNLRAFNDISGKGYKLMADVIMKVDKFNPMVATQLCDPFK LWNKLDLKRQALMHDEMNRMLSMDNISPNLKEYLLRLTNKM
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_1412200 | 539 S | IRPESYISM | 0.992 | unsp | PCHAS_1412200 | 539 S | IRPESYISM | 0.992 | unsp | PCHAS_1412200 | 539 S | IRPESYISM | 0.992 | unsp | PCHAS_1412200 | 669 S | VGLISPKDG | 0.998 | unsp | PCHAS_1412200 | 860 Y | KDESYVDFD | 0.991 | unsp | PCHAS_1412200 | 56 S | SQQHSSKIP | 0.991 | unsp | PCHAS_1412200 | 390 Y | SEEKYVSKL | 0.993 | unsp |