

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_1427150 | SP | 0.291110 | 0.702361 | 0.006529 | CS pos: 25-26. VLS-WC. Pr: 0.6374 |
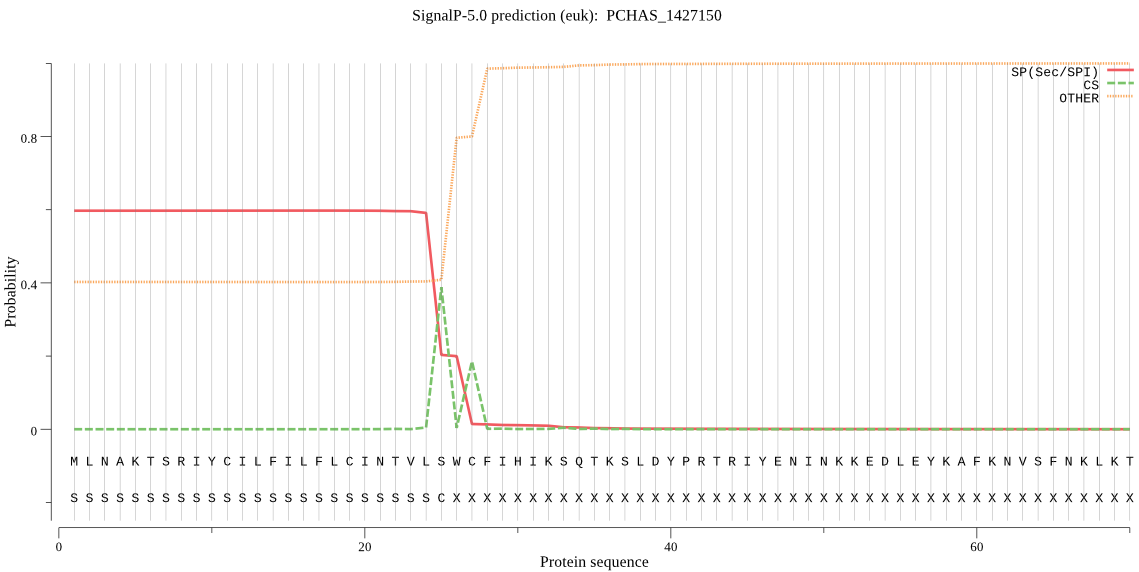
MLNAKTSRIYCILFILFLCINTVLSWCFIHIKSQTKSLDYPRTRIYENINKKEDLEYKAF KNVSFNKLKTSLKNFLVVNKKINISDVKYVSFLGKYYNYEKIDNYGVNEICILGRSNVGK STFLRNFIKYLINVNDNTNIKVSKRSGCTRSINLYSFENAKKKKLFILTDMPGLGYAEGI GEKKMEYLKKNLDDYVYLRNQICLFFILIDMSVDIQKIDISLVDAIRKTGIPLRVICTKC DKFNGDVNHRLNGIKTFYKLEKTPIHISKFSNHNYINIFKEIQYHCNLGNS
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PCHAS_1427150 | 143 S | NIKVSKRSG | 0.994 | unsp |