

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PCHAS_1463000 | SP | 0.000597 | 0.999394 | 0.000009 | CS pos: 19-20. AKG-DL. Pr: 0.9084 |
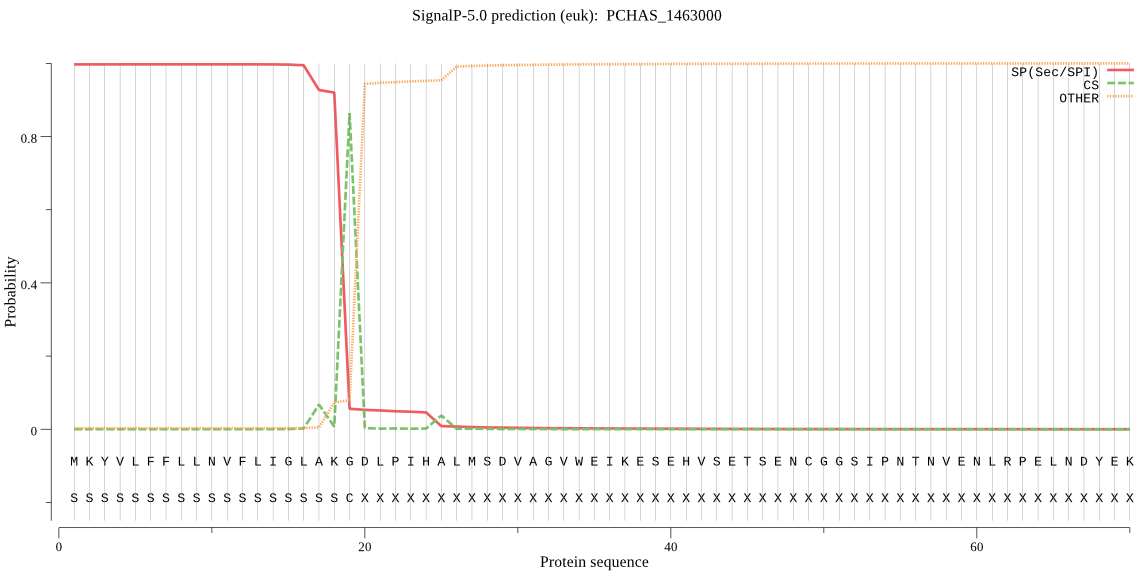
MKYVLFFLLNVFLIGLAKGDLPIHALMSDVAGVWEIKESEHVSETSENCGGSIPNTNVEN LRPELNDYEKYLQKNYGELKNFKIDLTLERIKLFNDDSSRGTWTYLAVRDVGHNNSVVGS WTMVYDEGFEIRLRGKRYFAFFKYDKKVSEECPSAVENINKINTECYKTDPTKIRLGWIL YERKKKNYEEKIYQWGCFYAEKIAKIPISSFVINNVKKYDKQDNKSVKNDDHAGSGTNFF VLMQRDNYSKIRMNHPELFSSTKISYIDKHPGKEDQIYGCRKRGSKKMEIDLVLPKEFSW GDPYNNNSFDEVVEDQKECGSCYLISSVYMLEKRFEILLSKRYKKNIKMNKLSHECIINL SKYNQACDGGFPFLVGKEIYENGICTDRRYGPIDSVSSGVEAFSKNKNNKIYYASDYNYI SGCYECSNEFDMMTEIINNGPIVVAVYATPLLLKVYELNDKNYIFTNISDENKICDVPNK GFNGWQQTNHAVAIVGWGEHTNENNELIKYWIIRNTWGSKWGYKGYLKYQRGINLNGIES QAVYIDPDFVRGSGKELIA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCHAS_1463000 | 265 S | STKISYIDK | 0.992 | unsp | PCHAS_1463000 | 265 S | STKISYIDK | 0.992 | unsp | PCHAS_1463000 | 265 S | STKISYIDK | 0.992 | unsp | PCHAS_1463000 | 285 S | RKRGSKKME | 0.995 | unsp | PCHAS_1463000 | 398 S | DSVSSGVEA | 0.994 | unsp | PCHAS_1463000 | 553 S | FVRGSGKEL | 0.997 | unsp | PCHAS_1463000 | 149 S | DKKVSEECP | 0.992 | unsp | PCHAS_1463000 | 226 S | QDNKSVKND | 0.997 | unsp |