

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PF3D7_0102400 | OTHER | 0.999980 | 0.000016 | 0.000004 |
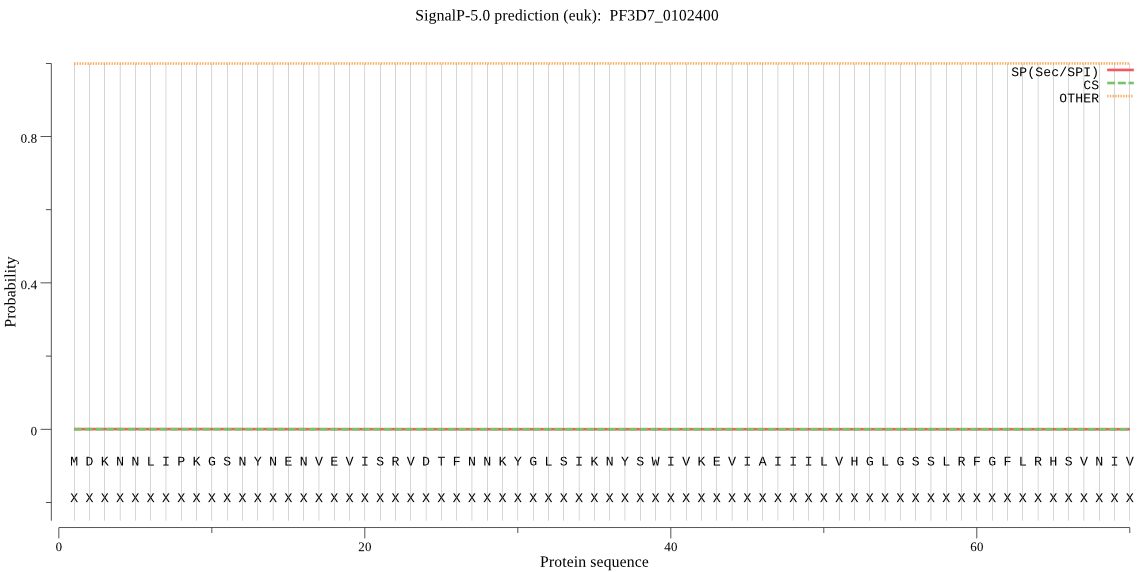
MDKNNLIPKGSNYNENVEVISRVDTFNNKYGLSIKNYSWIVKEVIAIIILVHGLGSSLRF GFLRHSVNIVDNMHATLIDSDNFYIYKNSWIEEFNKNGYSVYGIDLQGHGESDGIANLRL HINDFDDFSDDVIDHIKKIHHSIMFENDKKGNLYNNNNNNNNNNNNKMESMEKIPMYLIG YSMGGNIVLRTLEILGK*KDRISKYNIKGIICISAMVSVQLVGFTDSFKYRYFYLPATWL LATLFPTYRQKTGSVEFKKFPYVNDIISLDSGRFKGDKTNKFTYGIVKSLDTLHKYIDDI PRNIPILFIHSRNDSLCYYPGLEIFYNRLVSDNKELVTLENNEHLVIMEPGNEQILQKIL DWIFNIYKKEDERNNVKEEITLK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PF3D7_0102400 | 203 S | KDRISKYNI | 0.995 | unsp | PF3D7_0102400 | 254 S | QKTGSVEFK | 0.992 | unsp |