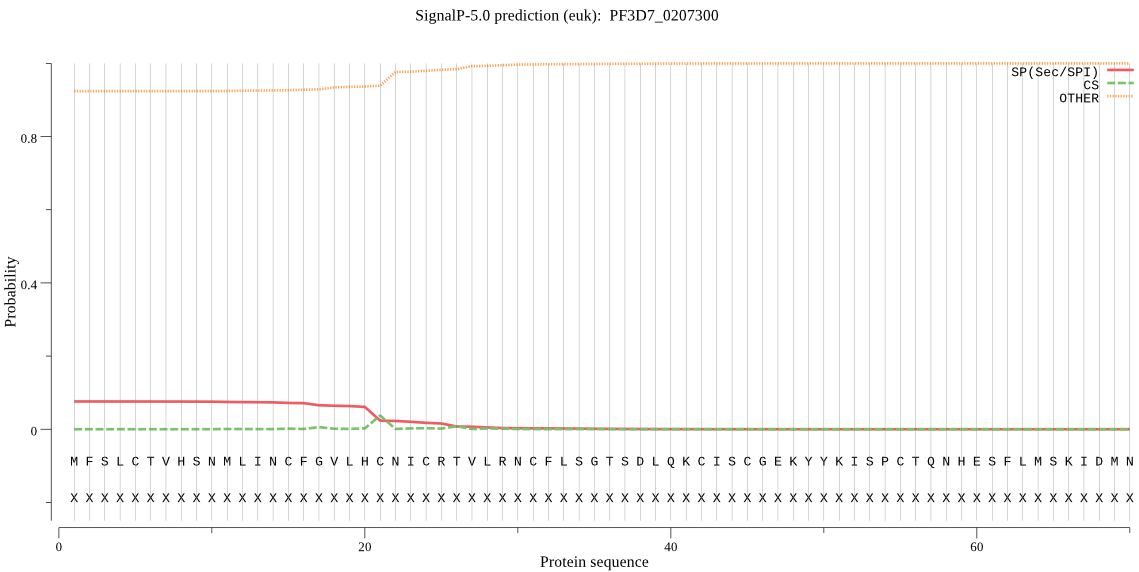
Fasta :-
MFSLCTVHSNMLINCFGVLHCNICRTVLRNCFLSGTSDLQKCISCGEKYYKISPCTQNHE
SFLMSKIDMNNTFVELKDHEEVTDEKMKNLITKIIEIAIDRHTLGLHDFSSVSDEYKEKI
KMLCMFSNYKDNYENANNHRQAKVEIVEEHIHKIVESYINEENNMEHMKDLLKNPALCLK
NPNQWVKDRAGFKDDDKPSVGIIPERKIFKPYDIKTLKSSLYASSTNCDRQFCDRFSDSN
ECEHRIRVLNQGKCGNCWVFASSVVIAAYRCRKGLGFAEPSIKYVTLCKNKHLMDIDNNP
FGHYNDNICKEGGHLSYYLETLEKTRMLPTSHDVPYNEPITGSECPDNKETWSNIWKGVN
LMDRIYAGYIYHGYFKVSFKDYVVSNRTNDLINIIKDYIIQQGSVFVSMEVTDKLTFDHD
GTKVMMSCEDNDSPDHALVLIGYGDYIKTNGKKSSYWLLRNSWGSHWGDKGNFKLDMYGP
NNCNGKVLYNAFPLLLNMAHNPIDVPLPNDLASTDIRVRYRQSDFNQNRNRNNYPQYDKN
SNDNDRNYINPYNKNDNNYNPYNKPHYNDKENDAYYEKNDDYNNAHIRRNTIRFKKRIIK
YSLYARIGNTVYKRTIFSKRKDEYKEPYSCLRTFSFEKDSDTKCRSNCEKYIDKCKHNSS
IGECLIQHSPNYKCVYCGM