

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
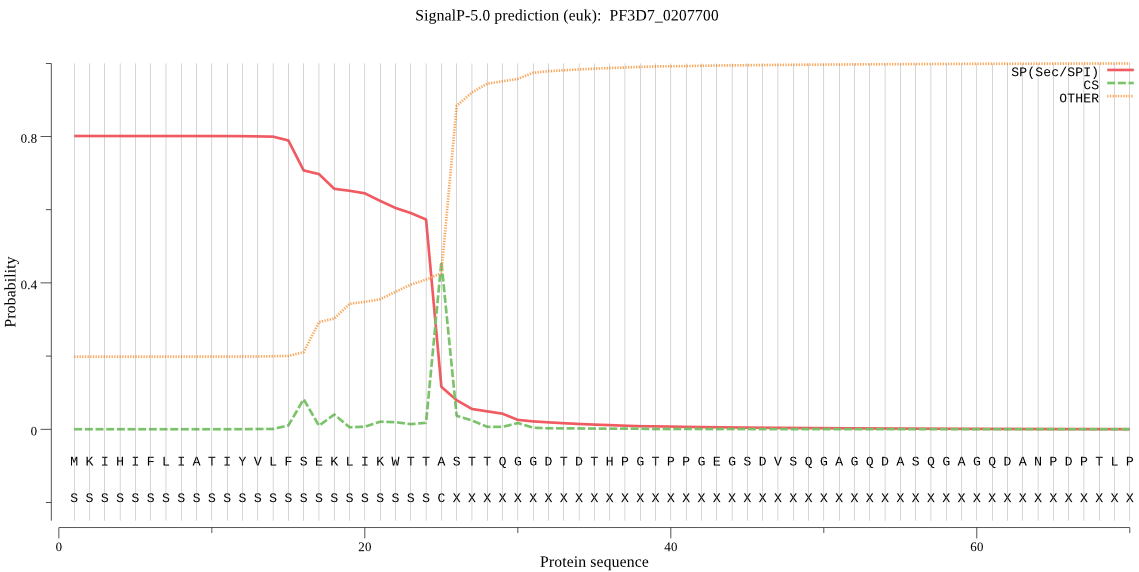
| PF3D7_0207700 | SP | 0.061882 | 0.937876 | 0.000242 | CS pos: 25-26. TTA-ST. Pr: 0.6583 |
MKIHIFLIATIYVLFSEKLIKWTTASTTQGGDTDTHPGTPPGEGSDVSQGAGQDASQGAG QDANPDPTLPKPPSPPADDTKDTGSQGDADSSSSKIEIPPLVKPENHKTIVSAMLKNYKG VKVTGTCGADFGLFLVPHIYVHVKSEDTEIELSSELAPPEMQTKFDKTQLKKFCVKDDTK KFDFIAYIYKDILVFKWKVYEEGLSKEQDVDEMKYLLPNLKQPITSIQVHSWTGTKESYI LESKDYVLGEGMPEKCDAIATDCFLSGFTDIGKCFQCKLLMQEKNINDSCFKYVSSNQKE LIKKQLKITAQDDEESSEYHLSESIKNLLKNIYKKNNDDNKKKELLHFENVNSALKSELL NYCNLLKEVNMNGVLKDHQLGNVQDVFNNLTKLLEEHKEENDNVLYHKMKNEALCLKNVN DWMKNKTGLLLPQLSYDLTYKNNNFTEFTQNKSYTSQNIVDKLYCNHEYCNRLKDHNNCI SKINVEDQKNCALSWAFASKYHLETIKCMKGYEPLNASVLYVTNCLKNKNKDVCTEGSNP LVFLETIEEKGFLPTESNYPYDQSKVGDICPQLQNDWDNVFENTKVLDYNNGPFSVGTKG YIAYESEAFQKDMHSFVKLVKDEIMNKGSVIAYVKAENVLGYELNGKKVQNLCGDKTPDH VVNIVGYGNYINNKGEKKSYWIVRNSWGKYWGDDGYFKVDMYGPSTCEDNFIHTVVVFNV QVPINEKFDKKEHDIYNYYLKTSPEFYHNLYYKTFNSNKEEKSMNKNSYVYGQDTTPVEN EAPRSGVQKPTELSSTESQTVSPPNESQTESLLSGGSQVTNPTLTQSTSSSSGQQETGPL STQGLSPATGDPKGKEQEASPAEGLSGVLNPTKEVTSEEKIQIIHLLKHIKNSKIRRGLV KYNHEFEVGDNSCSRSTSKNAEMHDECVNICEKYWPECRGTAVPGYCLSTHDDKNECDFC YV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PF3D7_0207700 | 877 S | KEVTSEEKI | 0.993 | unsp | PF3D7_0207700 | 877 S | KEVTSEEKI | 0.993 | unsp | PF3D7_0207700 | 877 S | KEVTSEEKI | 0.993 | unsp | PF3D7_0207700 | 85 S | KDTGSQGDA | 0.997 | unsp | PF3D7_0207700 | 94 S | DSSSSKIEI | 0.995 | unsp |