

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
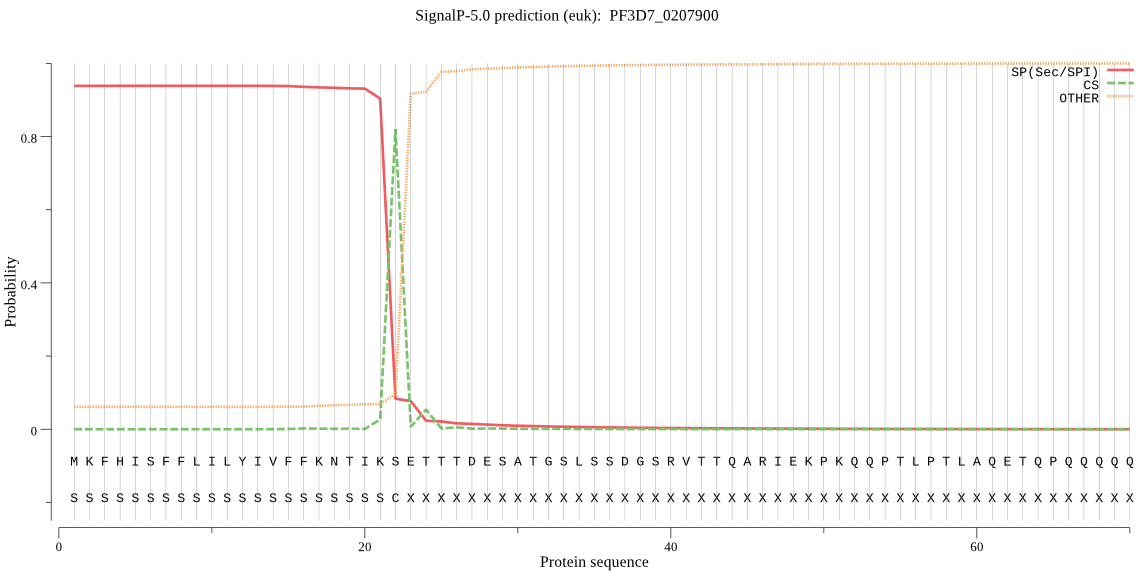
| PF3D7_0207900 | SP | 0.034154 | 0.965775 | 0.000071 | CS pos: 22-23. IKS-ET. Pr: 0.8590 |
MKFHISFFLILYIVFFKNTIKSETTTDESATGSLSSDGSRVTTQARIEKPKQQPTLPTLA QETQPQQQQQQKEVGSGIGAEQKVESARPGAEVSQSDVERAGRSSGTGGSVGTKISPGSQ GQGKVAGPQLPRLPQLPQSFEQSRNQQSSPVTPKRNGISPTNAKSPESVLPPAQSFTNLN KSTIPHTIPIKSSFLKYYKGVKITGSCGVQFQLVIVPHLFIYVETKENNIQLEPRFMKLN ERIDFEKDKSNLKNKCDVNKKQSFKFILYLQHDLITIKWKVYEEKPDTTTRIAPGQVDLN VDVKRYKLPKLDQPVISIQIHSLAQDGETYLMESKDYNLEDQIPEKCEAIASDCFLSGNV DIEKCLQCTLLVIKADKNDECLKYVSKNVKDRFEEILTKGEDDADSDEYDFIAPANYILK NIYKKNDTNGKKELLHFKDINNNLKLELINYCNLLKDNDVSGILTYEKLGNVQDIFNNLT KLLEEHKEENNYVLYHKMKNEVLCLKNANDWMKNKTGLVLPQLKYSLNKFNKNKENYIKE NIFEEDENGIVDLTKFPVDTSYSSYNYADSLYCNREYCNRLKDHNNCISKINVEDQKNCA LSWAFASIYHLETIKCMKGYEPLNASVLYVTNCLKNKNNDVCTEGSNPLVFLETIEEKGF LPTESNYPYDQSKVGDVCPQVQNDWDNVFENTKVLEYNNAPFSVGTKGYIAYESEVFQKD IDSFVKLIKDEIMNKGSVIAYVKAKNVLGYELNGKKVQNLCGDKKPDHAVNIVGYGNYIN NKGEKKSYWIVRNSWGKYWGDDGYFKVDMYGPPTCEDNFIHSVVVFNVEVPVNENFDKKE HDIYKSYLKNSPDFYHNIYYKNYNFENYVSPISTWSEVLHNTLYNNNLIWGQETTDPIGE GKLPASEAGHLRGEGKGNSEENQGQRSNANGAISSNQSTGGKITKQGESKDDGVALPVST DGKPNTSSSVVDTNDQRSLPNPRATSLQPPSVQIPNHEGTSAPGNSRTPSIVSPTAAEKS RKAQIFHVLKHIKNSRIKMGLVKYDNSDNIGGDHVCSRTYAVNPEKQEECVKFCEENWEN CKNKPSPGYCLAKLKNTNECFFCYV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PF3D7_0207900 | 969 S | NTSSSVVDT | 0.993 | unsp | PF3D7_0207900 | 969 S | NTSSSVVDT | 0.993 | unsp | PF3D7_0207900 | 969 S | NTSSSVVDT | 0.993 | unsp | PF3D7_0207900 | 1020 S | AAEKSRKAQ | 0.991 | unsp | PF3D7_0207900 | 105 S | AGRSSGTGG | 0.995 | unsp | PF3D7_0207900 | 949 S | KQGESKDDG | 0.997 | unsp |