

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PF3D7_0307400 | SP | 0.134125 | 0.864012 | 0.001863 | CS pos: 20-21. IQT-KK. Pr: 0.9358 |
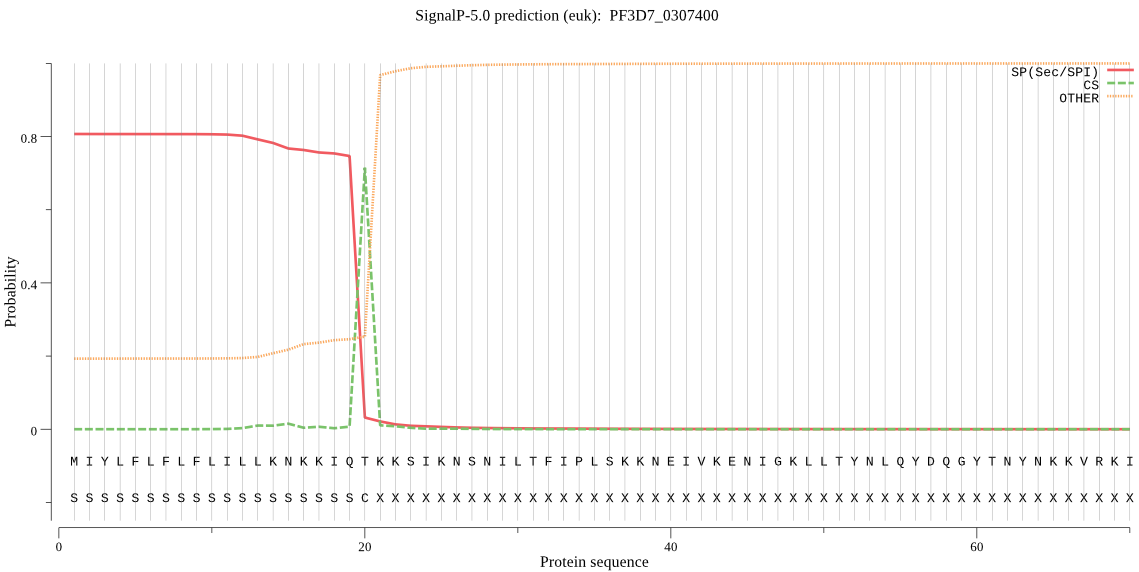
MIYLFLFLFLILLKNKKIQTKKSIKNSNILTFIPLSKKNEIVKENIGKLLTYNLQYDQGY TNYNKKVRKINVHNSSSEFANNINLDIEKQNNERAILKKEDLNFQIKNDCISKDNINKKK IKNMHNKDQGMIQHTNTNDNIDYTNSNYNNYLNYDVKKKKNKKRKYSKVQQQIITNNDDI KDMKKDVKLFFFKKRIIYLTDEINKKTADELISQLLYLDNINHNDIKIYINSPGGSINEG LAILDIFNYIKSDIQTISFGLVASMASVILASGKKGKRKSLPNCRIMIHQPLGNAFGHPQ DIEIQTKEILYLKKLLYHYLSSFTNQTVETIEKDSDRDYYMNALEAKQYGIIDEVIETKL PHPYFNKVEK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PF3D7_0307400 | 23 S | QTKKSIKNS | 0.994 | unsp | PF3D7_0307400 | 335 S | IEKDSDRDY | 0.998 | unsp |