

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
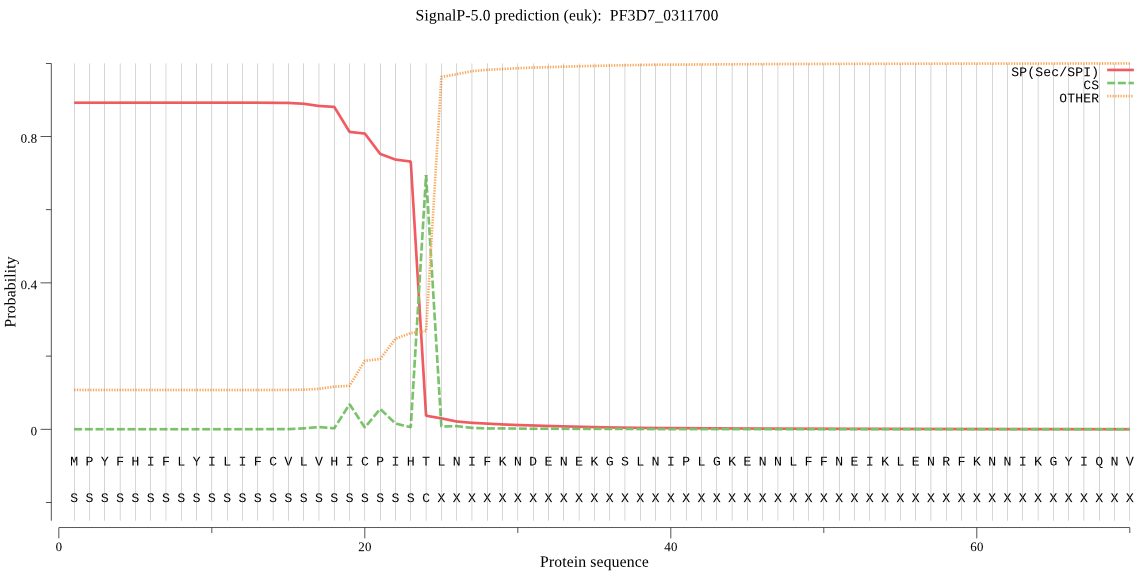
| PF3D7_0311700 | SP | 0.013282 | 0.986692 | 0.000026 | CS pos: 24-25. IHT-LN. Pr: 0.8758 |
MPYFHIFLYILIFCVLVHICPIHTLNIFKNDENEKGSLNIPLGKENNLFFNEIKLENRFK NNIKGYIQNVQKFHYLMEKNKPNVLSYIQEDLLNFHNSQFIADIGVGNPPQVFKVVFDTG SSNLAIPSTKCIKGGCASHKKFNPNKSRTFTKNLKNNQESVYTYIQYGTGTSILEQSYDD VYLKGLKIKHQCIGLAIEESLHPFSDLPFDGIVGLGFSDPDFRSQNKYASPLIETIKKQN LLKRNIFSFYVPKKLEKSGAITFGKANKKYTVEGKSIEWFPVISLYYWEINLLDIQLSHK NLFLCESKKCRAAIDTGSSLITGPSTFIQPLLEKINLERDCSNKESLPIISFVLKNVEGK EITLDFMPEDYIIEEGDTENNTLECVIGIMPLDVPPPRGPIFIFGNSFIRKYYTIFDNDH KLIGLIEANHNF
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PF3D7_0311700 | 276 S | VEGKSIEWF | 0.994 | unsp | PF3D7_0311700 | 342 S | ERDCSNKES | 0.994 | unsp |