

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PF3D7_0314300 | SP | 0.011616 | 0.988043 | 0.000342 | CS pos: 20-21. IHG-KD. Pr: 0.9554 |
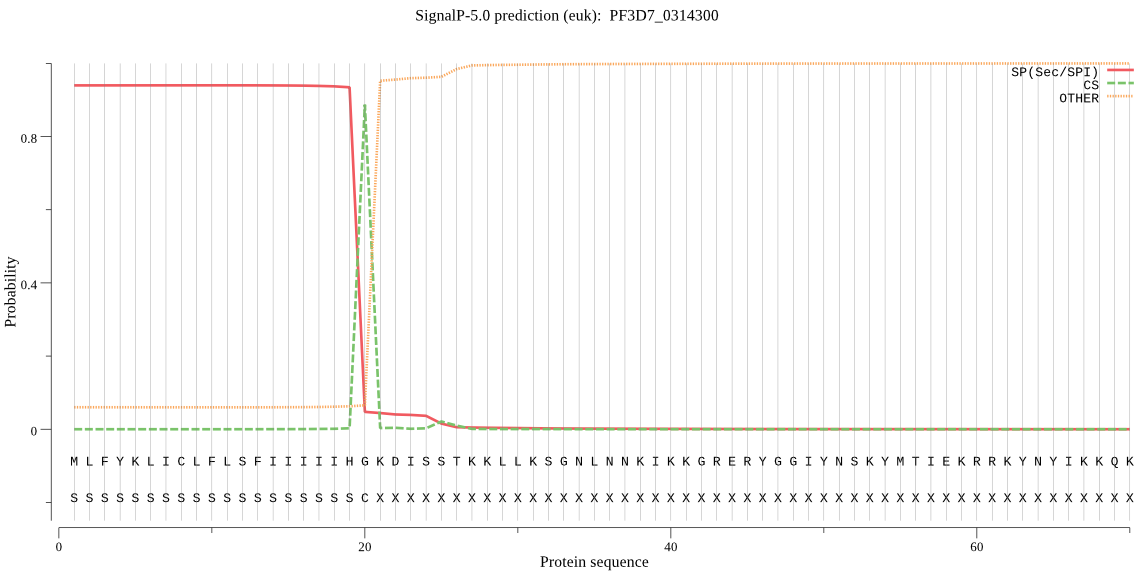
MLFYKLICLFLSFIIIIIHGKDISSTKKLLKSGNLNNKIKKGRERYGGIYNSKYMTIEKR RKYNYIKKQKNNNNSYFSYCNVYKNNDVNNNYTAYNYYINNPKNKLKEYYEKIKNHVIKK KKKIFSLKFSQNKRNEKKKKYFFINFTSFHDIKDNIKVLFNIDYIENNIIYSYIKSFKRT PPITKIYLLGTFLLSVLIHMNKNVYKLILFDFNKIFKKGEIWRLFTPYLYIGNLYLQYIL MFNYLNIYMSSVEISHYKKPEDFLIFLTFGYISNLLFTIWANMYNENIMNVKLYIHNFKN FFIKDCVSKYTSRSSTNNNSNNINSNNRSSNNNNHYNNSKNIDIKKEQYNHLGYVFSTYI LYYWSRINEGTLINCFELFFIKAEYVPFFFIIQNILLYNEFSLYEVASIFSSYLFFTYEK YFKFNYLRLFFKTLLKVIHIYPLYDRYKEEYE
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PF3D7_0314300 | 315 S | TSRSSTNNN | 0.992 | unsp |