

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
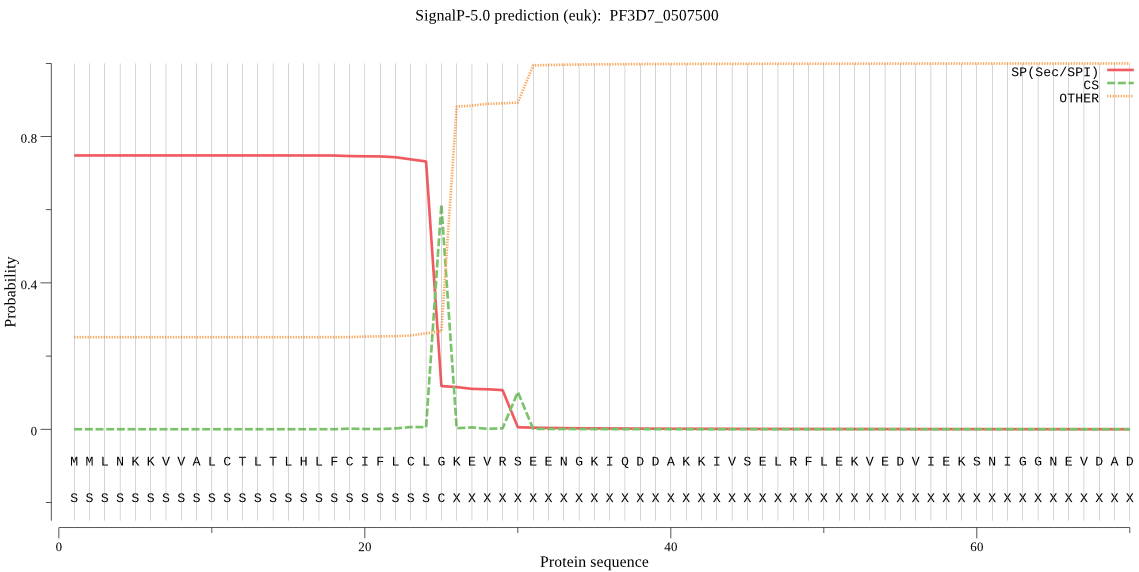
| PF3D7_0507500 | SP | 0.385818 | 0.613777 | 0.000406 | CS pos: 25-26. CLG-KE. Pr: 0.7104 |
MMLNKKVVALCTLTLHLFCIFLCLGKEVRSEENGKIQDDAKKIVSELRFLEKVEDVIEKS NIGGNEVDADENSFNPDTEVPIEEIEEIKMRELKDVKEEKNKNDNHNNNNNNISSSSSSS SNTFGEEKEEVSKKKKKLRLIVSENHATTPSFFQESLLEPDVLSFLESKGNLSNLKNINS MIIELKEDTTDDELISYIKILEEKGALIESDKLVSADNIDISGIKDAIRRGEENIDVNDY KSMLEVENDAEDYDKMFGMFNESHAATSKRKRHSTNERGYDTFSSPSYKTYSKSDYLYDD DNNNNNYYYSHSSNGHNSSSRNSSSSRSRPGKYHFNDEFRNLQWGLDLSRLDETQELINE HQVMSTRICVIDSGIDYNHPDLKDNIELNLKELHGRKGFDDDNNGIVDDIYGANFVNNSG NPMDDNYHGTHVSGIISAIGNNNIGVVGVDVNSKLIICKALDEHKLGRLGDMFKCLDYCI SRNAHMINGSFSFDEYSGIFNSSVEYLQRKGILFFVSASNCSHPKSSTPDIRKCDLSINA KYPPILSTVYDNVISVANLKKNDNNNHYSLSINSFYSNKYCQLAAPGTNIYSTAPHNSYR KLNGTSMAAPHVAAIASLIFSINPDLSYKKVIQILKDSIVYLPSLKNMVAWAGYADINKA VNLAIKSKKTYINSNISNKWKKKSRYLH
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PF3D7_0507500 | 132 S | KEEVSKKKK | 0.99 | unsp | PF3D7_0507500 | 132 S | KEEVSKKKK | 0.99 | unsp | PF3D7_0507500 | 132 S | KEEVSKKKK | 0.99 | unsp | PF3D7_0507500 | 274 S | RKRHSTNER | 0.997 | unsp | PF3D7_0507500 | 292 S | YKTYSKSDY | 0.993 | unsp | PF3D7_0507500 | 323 S | SSRNSSSSR | 0.993 | unsp | PF3D7_0507500 | 324 S | SRNSSSSRS | 0.997 | unsp | PF3D7_0507500 | 326 S | NSSSSRSRP | 0.995 | unsp | PF3D7_0507500 | 527 S | HPKSSTPDI | 0.995 | unsp | PF3D7_0507500 | 30 S | KEVRSEENG | 0.996 | unsp | PF3D7_0507500 | 119 S | SSSSSSSNT | 0.995 | unsp |