

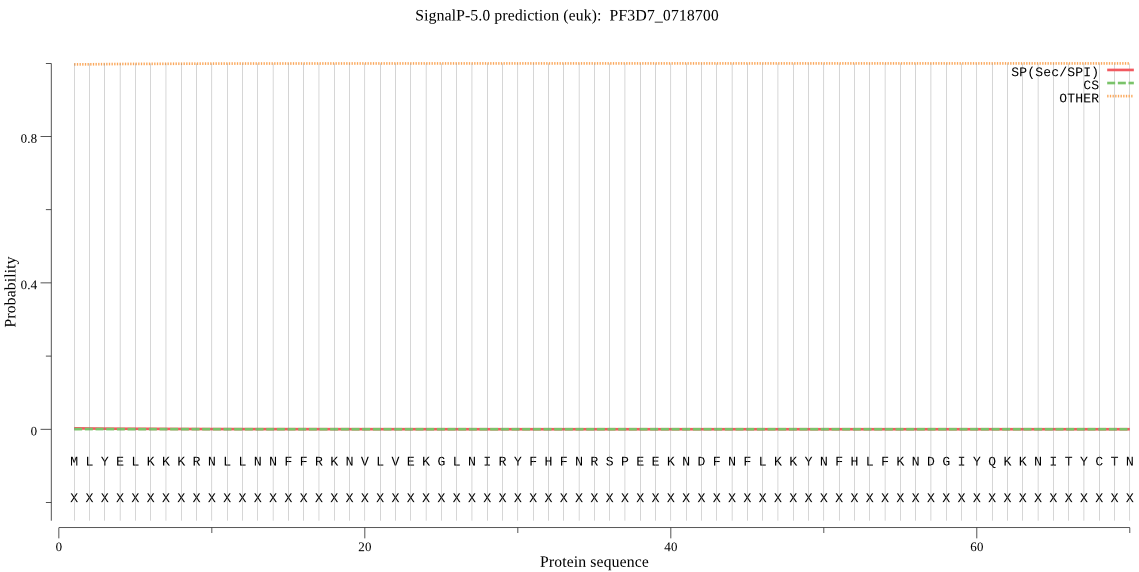
| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PF3D7_0718700 | OTHER | 0.960853 | 0.000038 | 0.039109 |
MLYELKKKRNLLNNFFRKNVLVEKGLNIRYFHFNRSPEEKNDFNFLKKYNFHLFKNDGIY QKKNITYCTNPKGQYKEFVKEKNNHYEKKEKVSDVLNYPESKSFPYASISIIIGMIGISG LLKFLIYNLKNCDNEKSIRLQVDKIIHHLDKYFILHNTEKRKGMDINNKIFNIKYITCAF YINENILQCSIQLLTFFLSSCFLEKYYGPIKYISLFLCGSIFSNLISLQFFKFLKQVESL RMVDFVLIHPSGSMAFICALCSLYFKKWAIWKNIPIHCSVLMVPYLISTFYGISSLYKIK KHNTINANKVTNDELKNYQKEMLQNNKGVLNNKSDHNDMINNINDNINIHNNTQEINKNP INMKDCRMVDTNNTLNTLKNYLIINACDSVIQKRKRENVFFNKKLQNLKKEAQECINEIN DKSEKMFFALSSAFTDMFGILLATAACCFMKCVKYIK
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PF3D7_0718700 | 36 S | HFNRSPEEK | 0.992 | unsp |