

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PF3D7_0803800 | OTHER | 0.991026 | 0.000547 | 0.008427 |
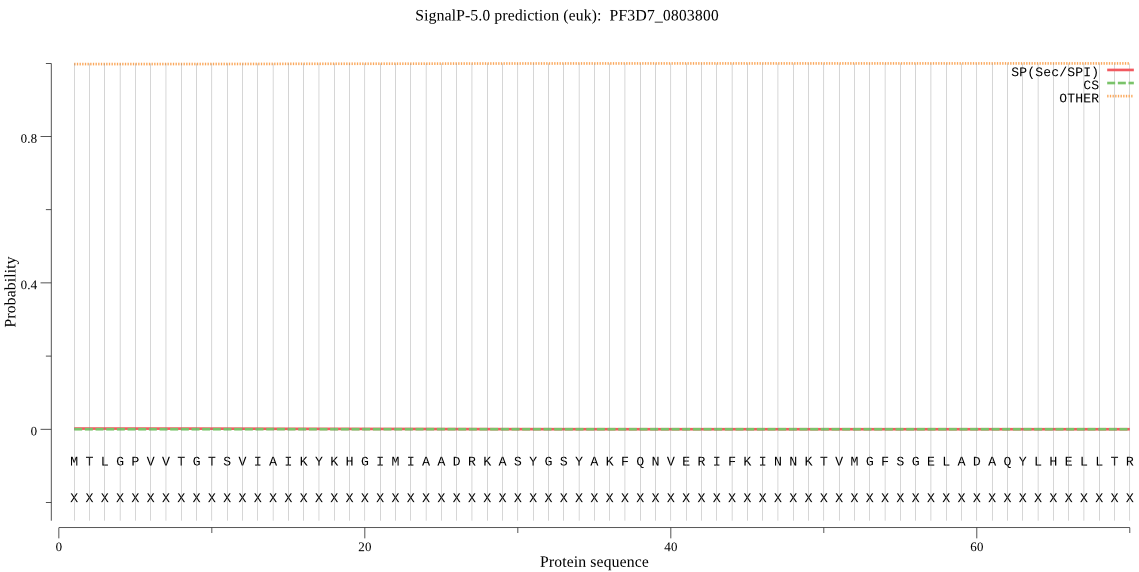
MTLGPVVTGTSVIAIKYKHGIMIAADRKASYGSYAKFQNVERIFKINNKTVMGFSGELAD AQYLHELLTRKNINNLSEKKRKEDMYTPQHYHSYVSRVFYVRKNRIDPLFNNIIIAGINS QKYDNNDDNVLLYTNKNNDDEQNEYKNNEEYKEIHKDDLYIGFVDMHGTNFCDDYITTGY ARYFALTLLRDHYKDNMTEEEARILINECLRILYFRDATSSNFIQIVKVTSKGVEYEEPY ILPCVLNSADYVYPSTLLPPAGCMW