

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PF3D7_0808200 | SP | 0.266724 | 0.729599 | 0.003677 | CS pos: 23-24. FKG-LK. Pr: 0.5213 |
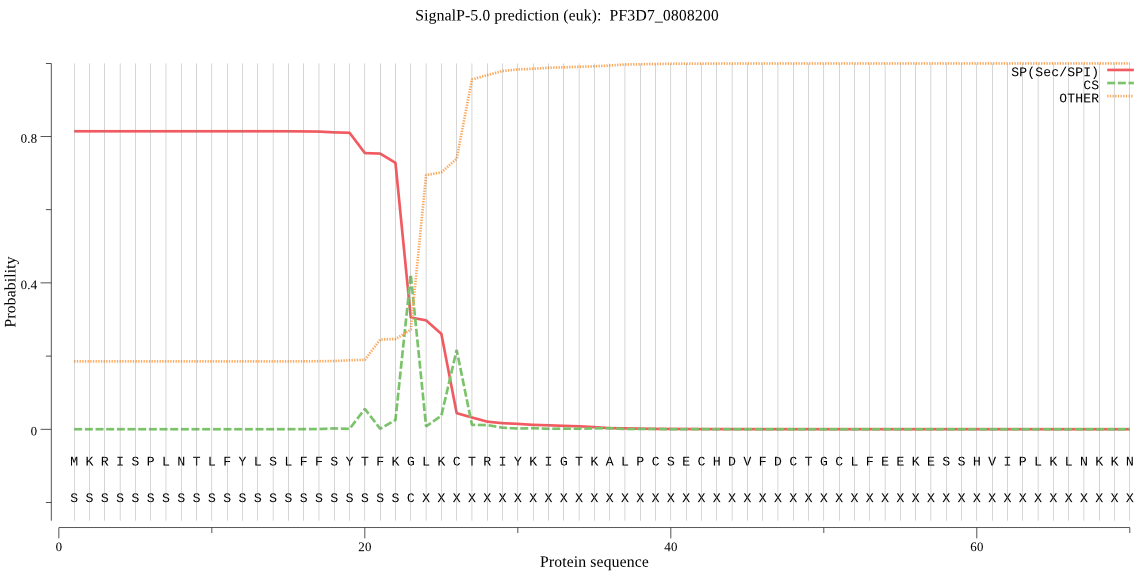
MKRISPLNTLFYLSLFFSYTFKGLKCTRIYKIGTKALPCSECHDVFDCTGCLFEEKESSH VIPLKLNKKNPNDHKKLQKHHESLKLGDVKYYVNRGEGISGSLGTSSGNTLDDMDLINEE INKKRTNAQLDEKNFLDFTTYNKNKAQDISDHLSDIQKHVYEQDAQKGNKNFTNNENNSD NENNSDNENNSDNENNLDNENNLDNENNSDNSSIEKNFIALENKNATVEQTKENIFLVPL KHLRDSQFVGELLVGTPPQTVYPIFDTGSTNVWVVTTACEEESCKKVRRYDPNKSKTFRR SFIEKNLHIVFGSGSISGSVGTDTFMLGKHLVRNQTFGLVESESNNNKNGGDNIFDYISF EGIVGLGFPGMLSAGNIPFFDNLLKQNPNVDPQFSFYISPYDGKSTLIIGGISKSFYEGD IYMLPVLKESYWEVKLDELYIGKERICCDEESYVIFDTGTSYNTMPSSQMKTFLNLIHST ACTEQNYKDILKSYPIIKYVFGELIIELHPEEYMILNDDVCMPAYMQIDVPSERNHAYLL GSLSFMRNFFTVFVRGTESRPSMVGVARAKSKN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PF3D7_0808200 | 453 Y | DEESYVIFD | 0.991 | unsp | PF3D7_0808200 | 453 Y | DEESYVIFD | 0.991 | unsp | PF3D7_0808200 | 453 Y | DEESYVIFD | 0.991 | unsp | PF3D7_0808200 | 562 S | ESRPSMVGV | 0.992 | unsp | PF3D7_0808200 | 5 S | MKRISPLNT | 0.991 | unsp | PF3D7_0808200 | 399 S | SFYISPYDG | 0.997 | unsp |