

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
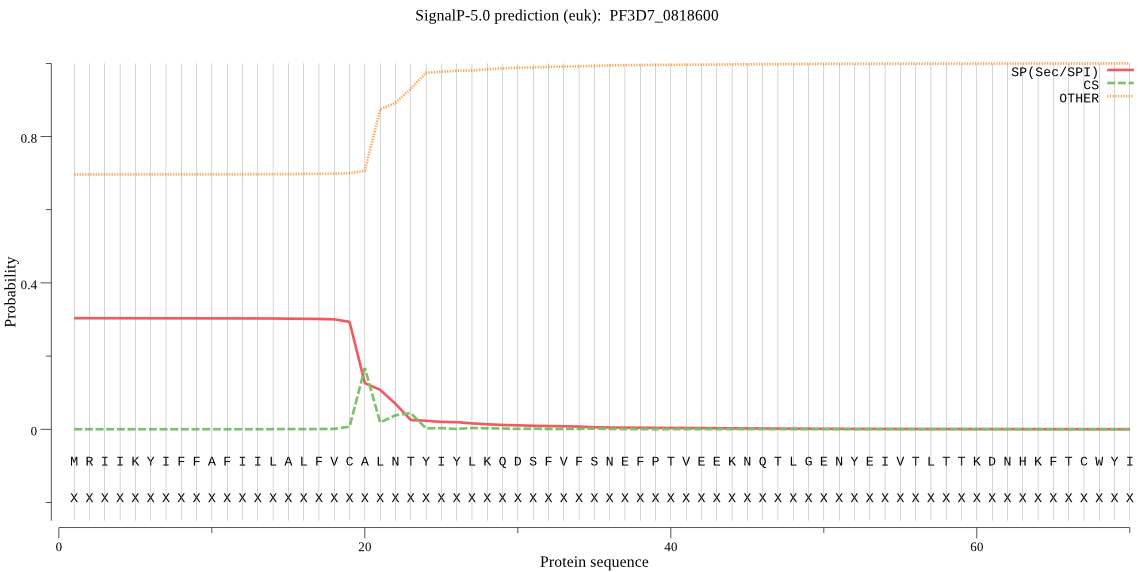
| PF3D7_0818600 | SP | 0.221414 | 0.778339 | 0.000247 | CS pos: 20-21. VCA-LN. Pr: 0.6327 |
MRIIKYIFFAFIILALFVCALNTYIYLKQDSFVFSNEFPTVEEKNQTLGENYEIVTLTTK DNHKFTCWYIKTKDSENKPIMLYFQGNGGYLEKYMNLFNLIIERVDVNIFSCSNRGCGSN IAKPSEEYFYKDAHVYIEYVKTKNPKHLFIFGSSMGAAVAIDTALKQHDHISGLIVQNAF TSLKELSRYSHPFLNYFLFDYDMIIRSKMDNETKIKNITVPTLFTLSEMDEKVPTSHTRT LFQLSASTNKQLYLSKGGTHPNILKNDDGSYHKAMKKFIVTAISIREKNIQKAVERNPNT PN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PF3D7_0818600 | 227 S | LFTLSEMDE | 0.992 | unsp | PF3D7_0818600 | 236 S | KVPTSHTRT | 0.994 | unsp |