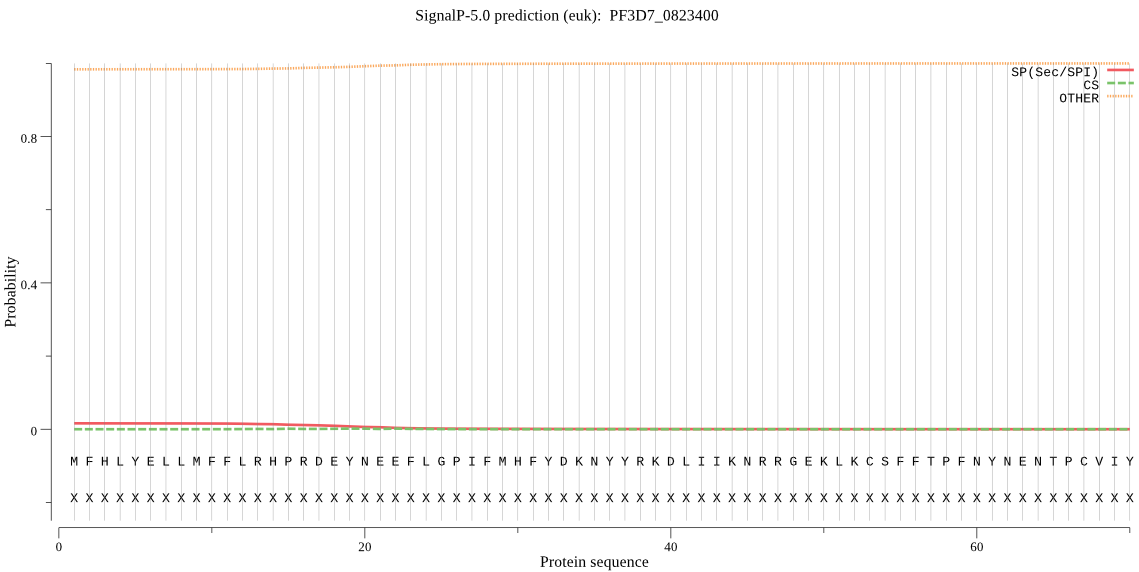
Fasta :-
MFHLYELLMFFLRHPRDEYNEEFLGPIFMHFYDKNYYRKDLIIKNRRGEKLKCSFFTPFN
YNENTPCVIYTHSASSCQLEVLDILHILLLCECSIFSYDCSGCGLSDGYYSTKGWNESQD
LYLLLNHLHYVEKIKNFVLWGKYSGAVSSIIAAALYGNIKLLILDSPYVSLIELYKTTFH
LNAKKKGEIFFKNVCLYLVRKQIKKKFHYDINNVCPIFFIEDITIPTIYIISKNDKIVHP
VHSLYFAYKQQKAYKIFYISESSTQTYENFSYENKLTLAIKTILYGCSINSTEFKKVFDV
YAYWKLFYSLKDKYENEFFFIDKLITKKLNQKNKIIKNVKKFILFKYESKTSLNSSTQST
MDSIREFNTNENEYSEQKLFTENKCENYKINQYKHSGNISECLYSDIDIMTDDHKVTSNP
FKDKHVDNLHNYNQTYGNIEEQQKESELLHMQSARSSLKSNKNTYKKSLTWDSKLQSSIT
YFKEDCPFQLLKND