

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PF3D7_0826300 | OTHER | 0.999963 | 0.000016 | 0.000022 |
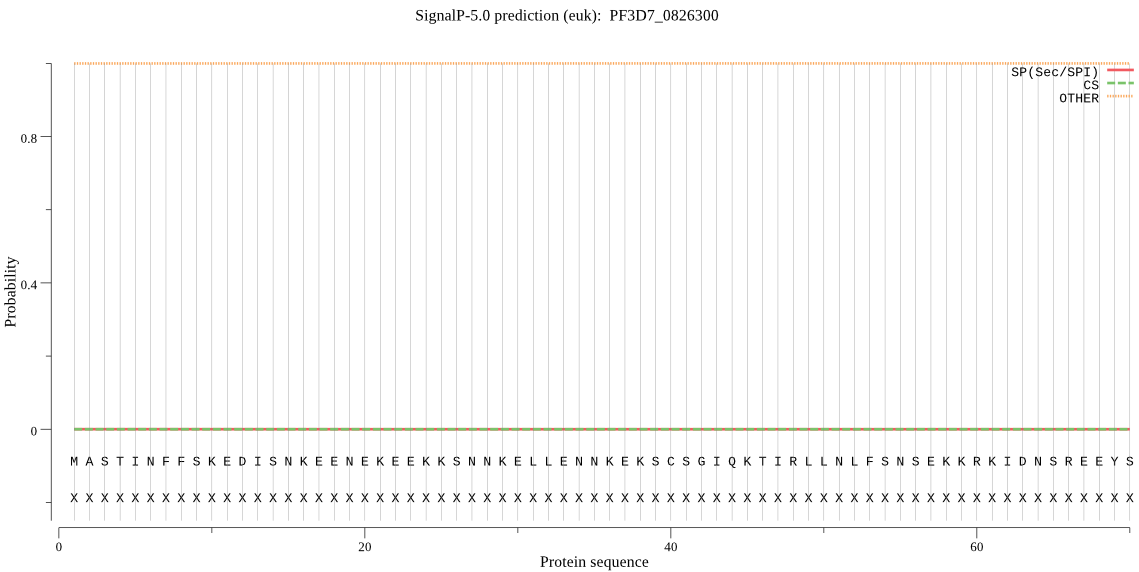
MASTINFFSKEDISNKEENEKEEKKSNNKELLENNKEKSCSGIQKTIRLLNLFSNSEKKR KIDNSREEYSDKILEKQSTLSLKDKEQNALNYINNEYVIEKGNDKLCEEQGQDHQISIAS LEDEGSYNKNVTTFERNESNNEKMKDIKNDDIKIKEKINYNKINNDRITYDGKSKKGEEK NKYSKEKTNTHLHVEVEKKDNIKHFFDYNKYRSKCNVTFSMKYKDSSVSLSNDKLTCYGD KGWSSVFVNNGADIGKWYYEIKIEEPVQNFNFLGYKDKIIKVNPYIRVGFACRYMRYDYP IGTDKYSYCVNSKNGKIFNNSISYDCMEPFNVGDIIGCYLNLKNKNSYNFDPRLDKKLYE HLQNGILCDPKNPPLLKKNEGSTIFFSLNGQIKKTSFMDIYEGFYHPSVSLYMGASAKIN LGPNFTYNHLQDYVPCVYMEPPTVL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PF3D7_0826300 | 65 S | KIDNSREEY | 0.997 | unsp | PF3D7_0826300 | 65 S | KIDNSREEY | 0.997 | unsp | PF3D7_0826300 | 65 S | KIDNSREEY | 0.997 | unsp | PF3D7_0826300 | 81 S | QSTLSLKDK | 0.998 | unsp | PF3D7_0826300 | 174 S | YDGKSKKGE | 0.997 | unsp | PF3D7_0826300 | 9 S | INFFSKEDI | 0.993 | unsp | PF3D7_0826300 | 14 S | KEDISNKEE | 0.993 | unsp |