

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
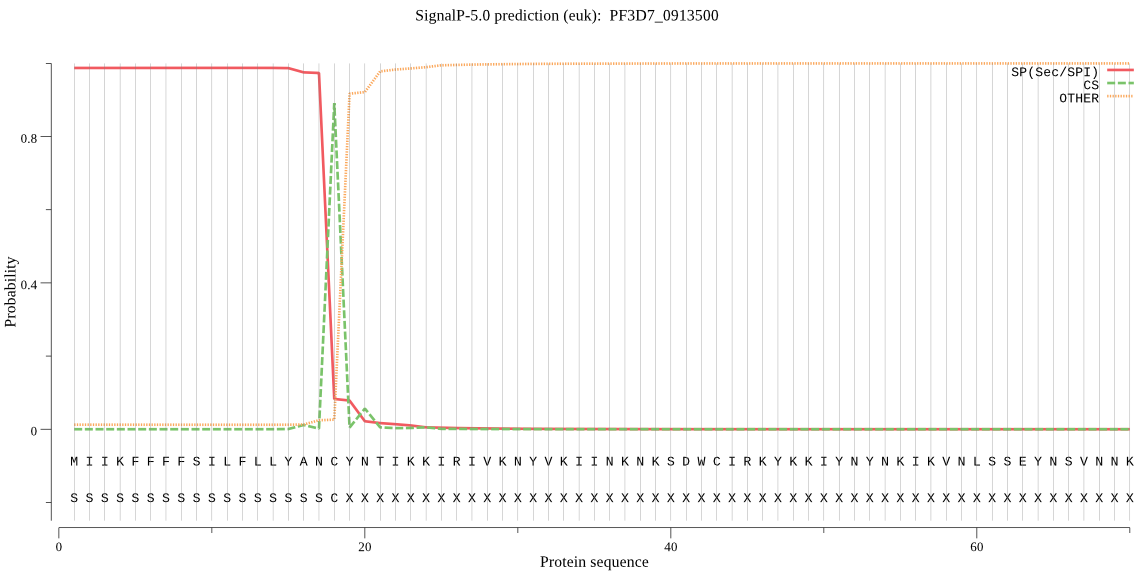
| PF3D7_0913500 | SP | 0.095672 | 0.903707 | 0.000622 | CS pos: 18-19. ANC-YN. Pr: 0.8627 |
MIIKFFFFSILFLLYANCYNTIKKIRIVKNYVKIINKNKSDWCIRKYKKIYNYNKIKVNL SSEYNSVNNKTNEQNNISKHIKELYIKNYIKIKNSFIHIKNILKNIKENLFQVTLKKQII ISVAFFFLHFYLLSEHFLILFPYQLIPNHSNILMSLDINNTLFLLTTLYFLKDIKTNFQT FRNKLKLNRFVLELQENKRKNILSISVLLIASYILSGYASIYTEKVLSILKLANITFNEN IIKSLQILAGHFIWVFCSYLTFRNLLYPYFKNNKSNLNFRYTDRWCFKVIYGYLFSHFIF NIVDIFNNFILNYFTSDDIYSDNSIDEIVHEKEFFSTFLCIISPCFSAPFFEEFIYRFFV LKSLNLFMNINYAVTFSSLFFAIHHLNIFNLIPLFFLSFFWSYIYIYTDNILVTMLIHSF WNIYVFLTSLYN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PF3D7_0913500 | 320 Y | SDDIYSDNS | 0.991 | unsp | PF3D7_0913500 | 324 S | YSDNSIDEI | 0.992 | unsp |