

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PF3D7_1011400 | OTHER | 0.986663 | 0.013330 | 0.000007 |
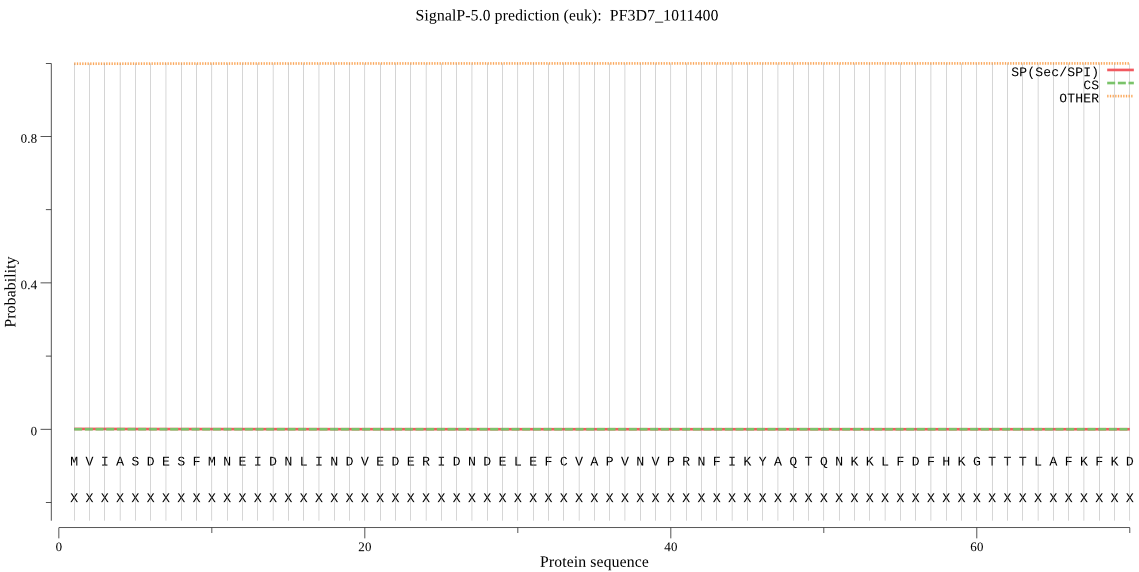
MVIASDESFMNEIDNLINDVEDERIDNDELEFCVAPVNVPRNFIKYAQTQNKKLFDFHKG TTTLAFKFKDGIIVAVDSRASMGSFISSQNVEKIIEINKNILGTMAGGAADCLYWEKYLG KIIKIYELRNNEKISVRAASTILSNILYQYKGYGLCCGIILSGYDHTGFNMFYVDDSGKK VEGNLFSCGSGSTYAYSILDSAYDYNLNLDQAVELARNAIYHATFRDGGSGGKVRVFHIH KNGYDKIIEGEDVFDLHYHYTNPEQKDQYVM