

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
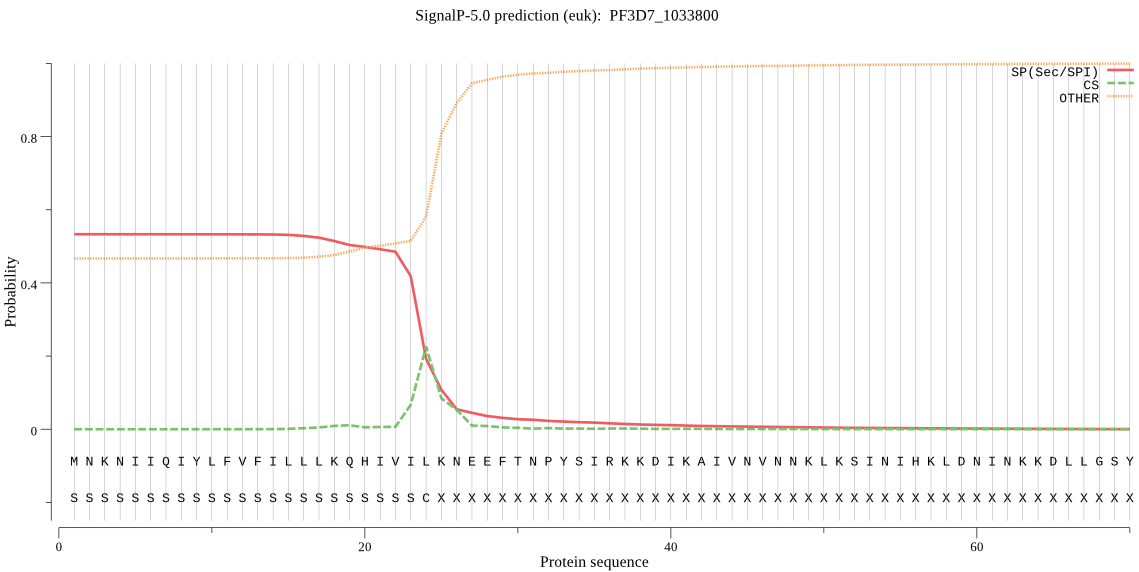
| PF3D7_1033800 | SP | 0.204828 | 0.789352 | 0.005820 | CS pos: 24-25. VIL-KN. Pr: 0.4894 |
MNKNIIQIYLFVFILLLKQHIVILKNEEFTNPYSIRKKDIKAIVNVNNKLKSINIHKLDN INKKDLLGSYNENYILIKLKKQDIFSKKLSTYYGEVQIGEQSENNMNVLFDTGSSQVWIL NDTCKNSLCNNIHSKYKRTKSFVYKYDKKGLPSVIEIFYLSGKIVAFEGYDTIYLGKKLK IPHTNISFATKVDIPILEEFKWDGIIGLGFQNGDSIKRGIKPFLDILKDDKILTNKNYKN QFGYYLSDKEGYITLGGIDNRLKNTPDEEIIWTPVSTEMGYWTIQIMGIRKEYVNNHFEE NKEEEEVIVKYEAFHDGGKNSIIDTGTYLIYAPKNTMENYLKDLKINNCDEKYNLPHLIF QIKSDEIKTIKGSAIIEIVLTPNDYVIEYVDKKNNTKECILGIQPDEQSEEDNVDGWTLG QVFLKAYYTIFDKDNLKIGFVRSKRNVTLR
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PF3D7_1033800 | 215 S | QNGDSIKRG | 0.992 | unsp | PF3D7_1033800 | 409 S | PDEQSEEDN | 0.995 | unsp |