

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PF3D7_1128700 | SP | 0.010111 | 0.989695 | 0.000193 | CS pos: 20-21. VCG-SV. Pr: 0.9363 |
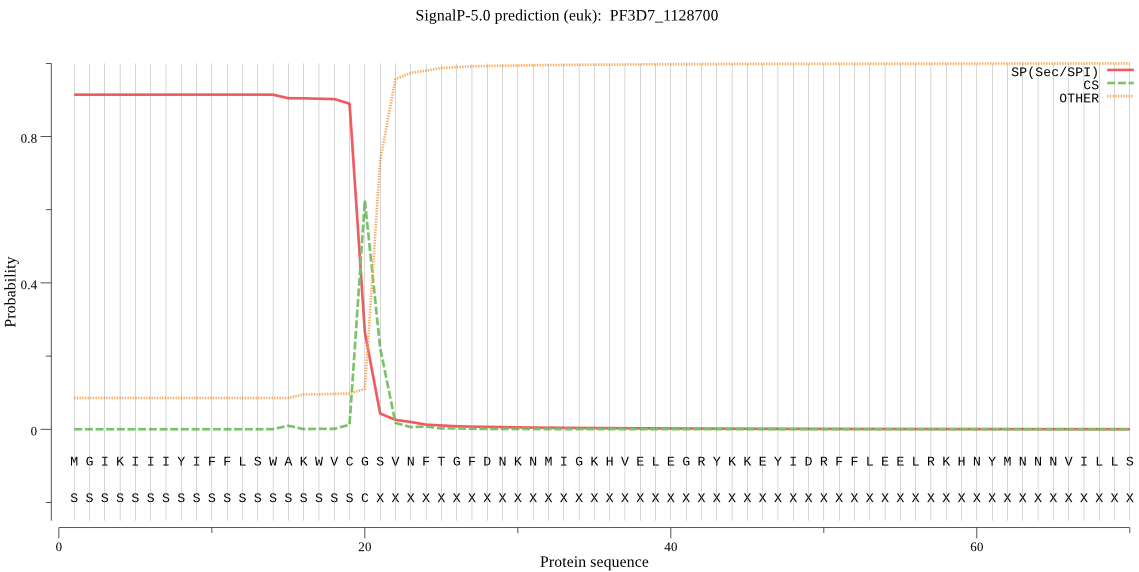
MGIKIIIYIFFLSWAKWVCGSVNFTGFDNKNMIGKHVELEGRYKKEYIDRFFLEELRKHN YMNNNVILLSTSRHYFNYRHTTNLLIAYKYLKYFGDTMDKNILLMIPFDQACDCRNIREG QIFREYELFPSSHNKETKIENINLYENLNIDYKNNNVRDEQIRRVLRHRYDAFTPKKNRL YNNGNNEKNLFLYMTGHGGVNFLKIQEFNIISSSEFNIYIQELLIKNFYKYIFVIIDTCQ GYSFYDDILNFVYKKKINNIFFLSSSKRNENSYSLFSSSYLSVSTVDRFTYHFFNYLQQI HKIYEKEPSKNIKAFSLYNILNYLKTQHIMSEPTTNNSKFNSSIFLHDKNILFFNSNLLI IHKDDVSIIYQDKQTHNHKYICLDNLSKCGHIKNNVHKKMQTLYEQTLYYNNNQQNFFSN HMSNFTDYFFTHDIYNIYNIYNVYNIYNVYNIYNVYDIYNVYSFLILLLSLFFIMCSLLT YYIVFFTEKAKMT