

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PF3D7_1143000 | SP | 0.097450 | 0.901657 | 0.000893 | CS pos: 19-20. NLS-ER. Pr: 0.4872 |
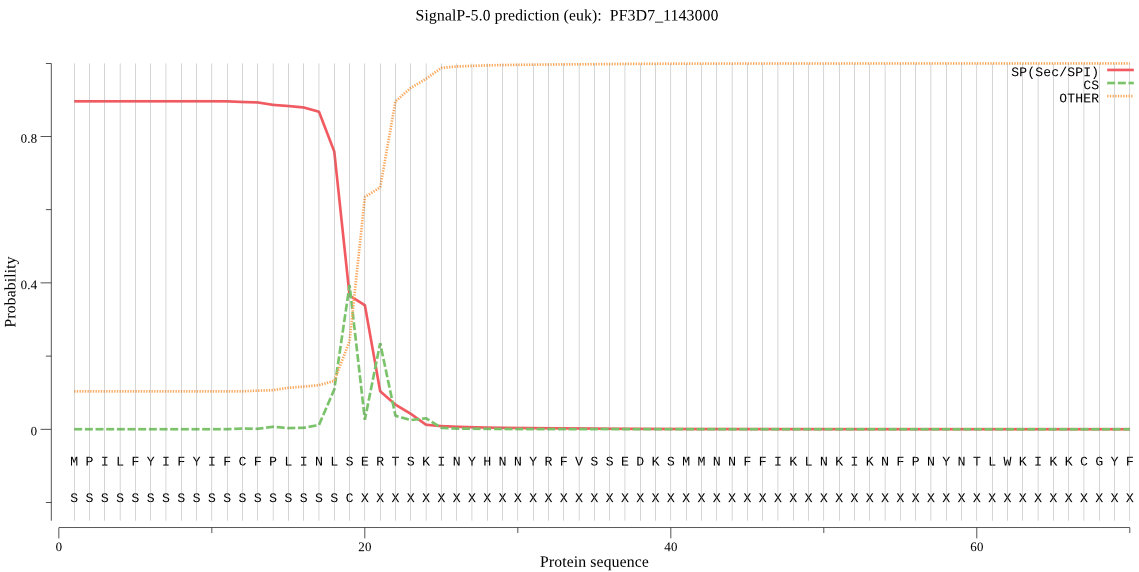
MPILFYIFYIFCFPLINLSERTSKINYHNNYRFVSSEDKSMMNNFFIKLNKIKNFPNYNT LWKIKKCGYFFLSYKNGKNNWNRISVNTITSSNNNEKITDDVLDGISFSIRDNTHIFKNE TKDIPIVLLHGCYGSRKNFIFFSKLLKSNKVITMDLRNHGDSKHTENMRFDEIENDIKNV LKKLHIKECCLIGFSLGGKASMYCALKNSSLFSHLIIMDILPFNYNCNKNPIKLPFNISQ VTSILYHIKHEKKPRNKLEFLQYLKCELPDISNSFAQFLCMSLKENNDKNQLTWKINIDA IYKDLPFIMNFPLNSQEYKYLNPCNFIIAKKSDLVCSIPNFDKIIKDYFPSASQIILENS THTVYIDEAQQCADIINGMLNK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PF3D7_1143000 | 35 S | YRFVSSEDK | 0.996 | unsp | PF3D7_1143000 | 109 S | GISFSIRDN | 0.991 | unsp |