

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
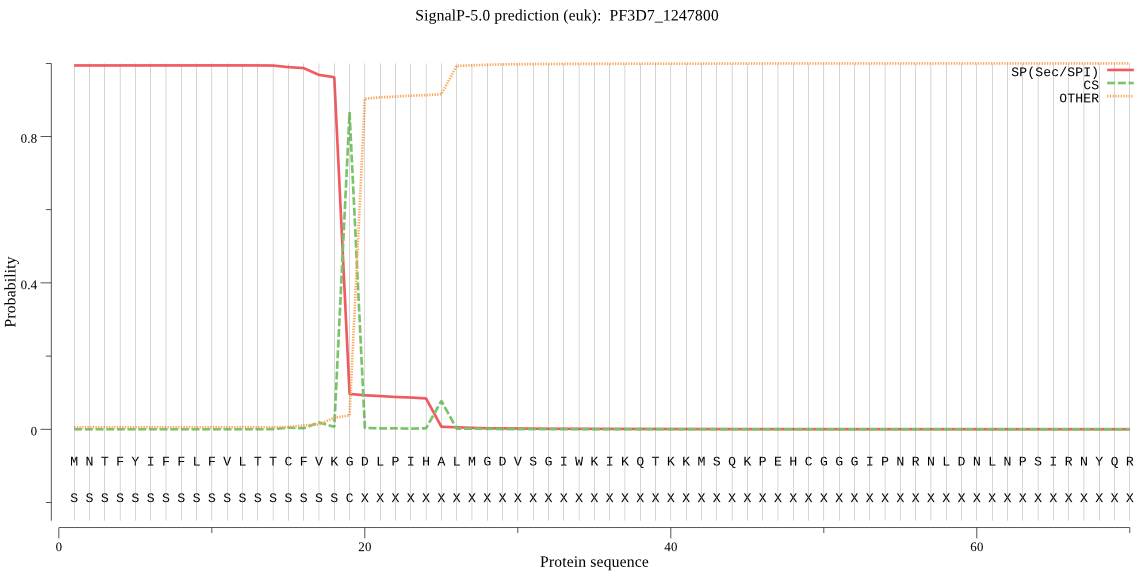
| PF3D7_1247800 | SP | 0.001124 | 0.998846 | 0.000030 | CS pos: 19-20. VKG-DL. Pr: 0.8295 |
MNTFYIFFLFVLTTCFVKGDLPIHALMGDVSGIWKIKQTKKMSQKPEHCGGGIPNRNLDN LNPSIRNYQRFLENEYGNLDMMIVNLTMEKVKIINQEKPRDKWTYLAVRDYERNEIIGHW TMVYDEGFEIRLNGSKYFAFFKYERKSNAHCPTSIEDKSYNDRDCYKTNPTQTHIGWVLN EKVKENTKEKIFYWGCFYAEKKESTPVSSFVLHNGMENKTNLVESHDYHFEEKKKKNLPP IGRRSNYKKFRFSNLQKIYGSTQTHLRDIYGRSGERKYGCRKRDILNLKIRLTLPKQFSW GDPFNDENFEENVDDQKDCGSCYSISSVYSLERRFEILFWKKYKKKVNMPRLSHQSILSC SPYNQGCDGGYPFLVGKHMYEYGIIPEQYMHYENNDYNNCIMDMGNYNHLNKQNRNIKEI YYVSDYNYINGCYECTNEYEMMNEIILNGPIVAAINATSELLNFYNIEDKNVVYDILPKD THQVCDVPNKGFNGWQQTNHAINIVGWGEQIINQDKIKNDDANNNNNDNNHKLVKYWIIR NTWGKNWGYKGYLKFQRGINLAGIESQAVYIDPDFSRGYPKNILQSDSLE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PF3D7_1247800 | 299 S | PKQFSWGDP | 0.994 | unsp | PF3D7_1247800 | 299 S | PKQFSWGDP | 0.994 | unsp | PF3D7_1247800 | 299 S | PKQFSWGDP | 0.994 | unsp | PF3D7_1247800 | 154 S | HCPTSIEDK | 0.996 | unsp | PF3D7_1247800 | 159 S | IEDKSYNDR | 0.995 | unsp |