

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PF3D7_1358300 | SP | 0.242905 | 0.753961 | 0.003134 | CS pos: 23-24. GNC-FI. Pr: 0.9801 |
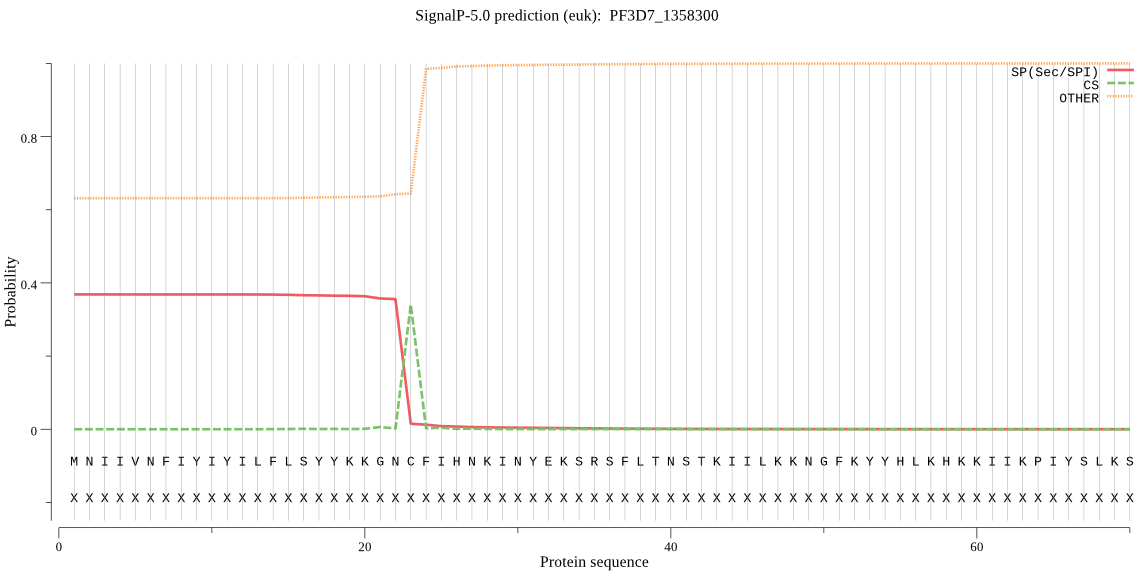
MNIIVNFIYIYILFLSYYKKGNCFIHNKINYEKSRSFLTNSTKIILKKNGFKYYHLKHKK IIKPIYSLKSSINNIWNGIKSIKVKKLLGIFKSEELKRHFHNYNLSYASYIFNKCRLDRV LISINVLLYLYLNRIDKNEERKIYFHKGNLIQEQDEQKCEKYKCNYNDVYKNRNYKTFLT SIFIHKNILHLYFNMSSLISIYRLISNVYTNTQIFLIFLLSGFFSNLISYYNHFKKKNEE IYLNDIIDQNYINKKIFNFKNNKIICGSSSAIYALYGMHITHVIFFYFKNHYVINTSFLY NFFYSFISSLLLENVSHFNHIVGFLCGFVFSFLIIAFEKN