

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PF3D7_1436800 | SP | 0.012379 | 0.979950 | 0.007671 | CS pos: 17-18. CKC-TK. Pr: 0.9484 |
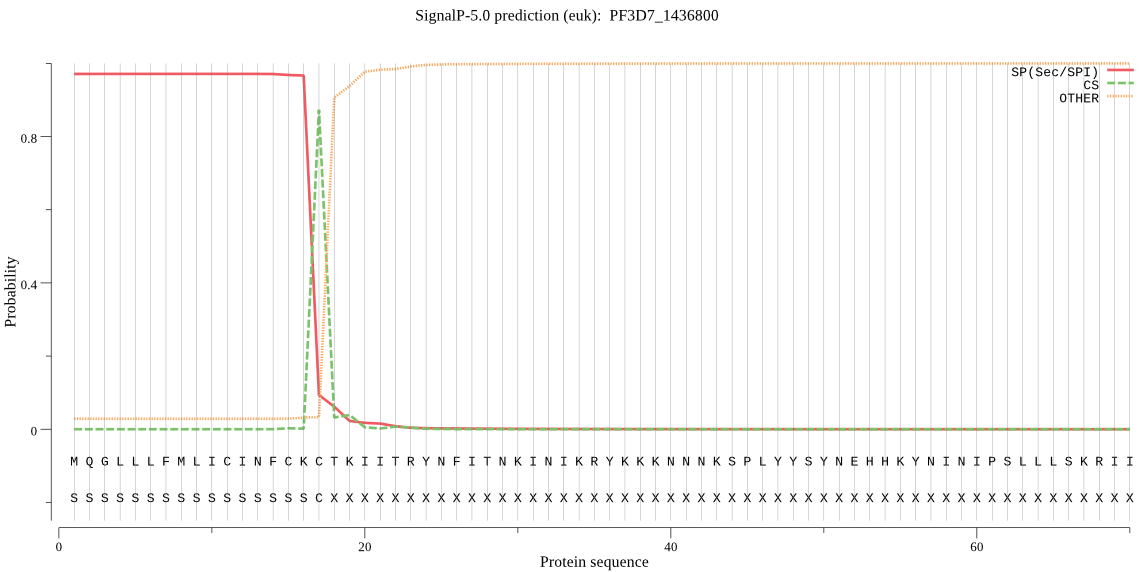
MQGLLLFMLICINFCKCTKIITRYNFITNKINIKRYKKKNNNKSPLYYSYNEHHKYNINI PSLLLSKRIIFLSSPIYPHISEQIISQLLYLEYESKRKPIHLYINSTGDIDNNKIINLNG ITDVISIVDVINYISSDVYTYCLGKAYGIACILASSGKKGYRFSLKNSSFCLNQSYSIIP FNQATNIEIQNKEIMNTKKKVIEIISKNTEKDTNVISNVLERDKYFNADEAVDFKLIDHI LEKE