

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PF3D7_1452300 | SP | 0.035380 | 0.963509 | 0.001111 | CS pos: 19-20. IHC-KN. Pr: 0.9873 |
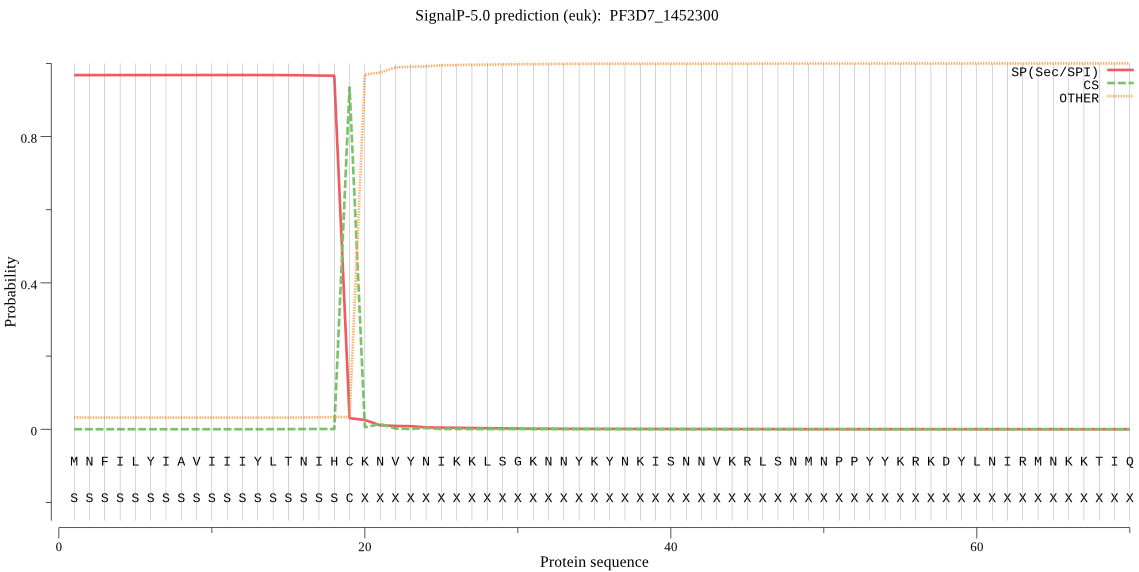
MNFILYIAVIIIYLTNIHCKNVYNIKKLSGKNNYKYNKISNNVKRLSNMNPPYYKRKDYL NIRMNKKTIQNFHTTNQNIHYKKFKNFVNKSKNKNKLYYLYIPNNDTNSIKGKKNKQNNK VLEIYFQQKKLLAQYFNNLKSSFIYKLKNTKIITKLFLSSSLLILTLNVIGLKPEDIALH SKRVLRAFEFYRIYTSALFYGDISLYVLTNIYMLYVQSNQLENILGSSEMLSYYISQISI LSIICSYIKKPFYSTALLKSLLFVNCMLNPYQKANLIFGININNMYLPYLSILIDIIHAQ NLKASISGLLGVTSGSIYYLLNIYLYDNYNKKVFKIPKFLETYLDSYDTIEIIQ