

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0104300 | SP | 0.059325 | 0.937894 | 0.002781 | CS pos: 26-27. CKA-VA. Pr: 0.4805 |
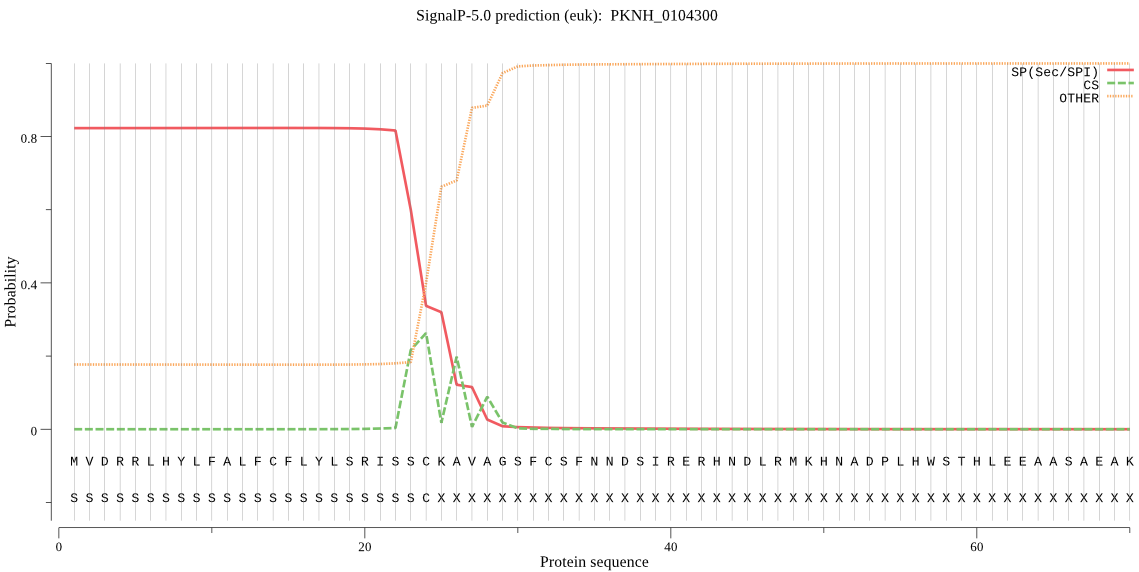
MVDRRLHYLFALFCFLYLSRISSCKAVAGSFCSFNNDSIRERHNDLRMKHNADPLHWSTH LEEAASAEAKLIKDMSNCTVLVNQINTNYFTISTKSKIESAVDTWYEGINNYDFELGPIR RGDDTVFEFTRVVWNSAEHIGCSSACCGNRGVLICKYDNNTNQPGHFADNVGTLDPMFVW ENFAFAPEQRRPGSETSQNDLPPLPIS
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PKNH_0104300 | 96 S | ISTKSKIES | 0.996 | unsp |