

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
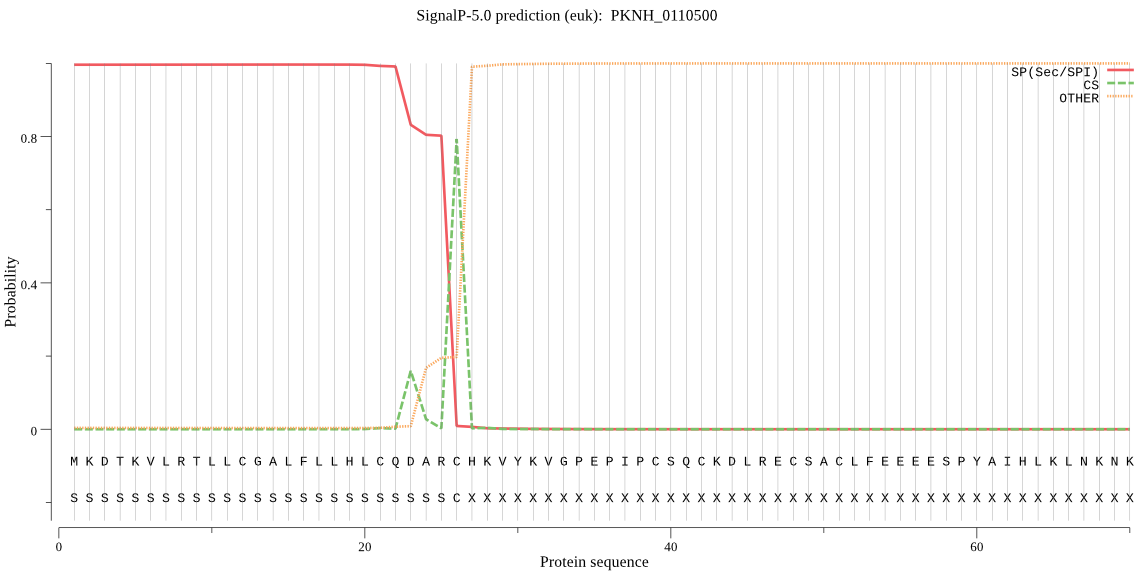
| PKNH_0110500 | SP | 0.014989 | 0.984960 | 0.000051 | CS pos: 26-27. ARC-HK. Pr: 0.9011 |
MKDTKVLRTLLCGALFLLHLCQDARCHKVYKVGPEPIPCSQCKDLRECSACLFEEEESPY AIHLKLNKNKPNDHSNLKKHHDSLKLGGVKYYVKRGEGISGSLGNPLGNTMDDIDSINEE IQNRRKESAGGGRNFIEMSNYKKDSLSDYFSGVQKHAHSEVGRVNMGDEKGRRENEGGEG GEGHASEHADRVERNFIDLKNTNAVVEQTEENVFLIPLKHLRDSQFVGKLLVGVPPQEIH PIFDTGSTNLWVVTTDCEEKSCKKVQRYNPYKSKTFRRSFIGKNLHIVFGSGSISGSIGK ETFVLGNHTVRNQTFGLVESESNDSLNGDNIFDYIDFEGIVGLGFPEMLSAGKVSFFDNL LKQNKNLSPQFSFYISPDDNTSTFIIGGLSKSFYQGSIYMLPVIKEYYWEVELDGIYVGE KKICCEEKSYAIFDTGTSYNTMPSAQIKNFFDVVPSVACTEENYQDVLKNYPIIKYVFGD LIIELMPEEYMILNEDNCIPAYMQIDVPSEKNHAYLLGSLAFMRHYYTVFVRGVNGKPSM VGVAKAKSAASAVN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0110500 | 145 S | YKKDSLSDY | 0.995 | unsp | PKNH_0110500 | 376 S | SFYISPDDN | 0.995 | unsp |