

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
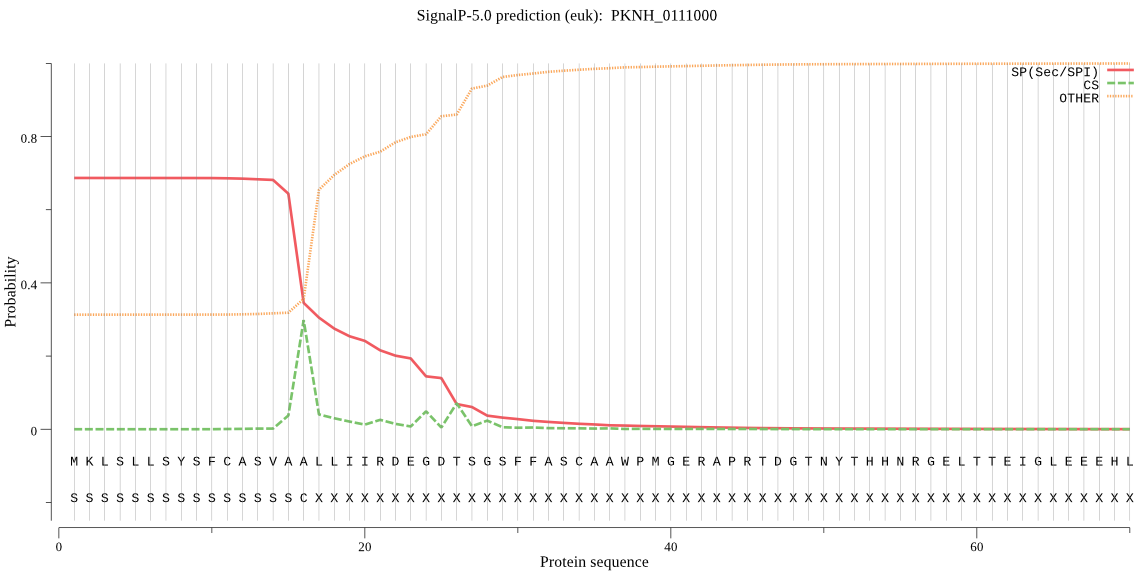
| PKNH_0111000 | SP | 0.030604 | 0.968619 | 0.000777 | CS pos: 16-17. VAA-LL. Pr: 0.6354 |
MKLSLLSYSFCASVAALLIIRDEGDTSGSFFASCAAWPMGERAPRTDGTNYTHHNRGELT TEIGLEEEHLYNDTETQGGDDSELVFNKRTPSVRSKSSTVDPPFVKYVPAYVNLPVNYKP VAKMNAAGGEEESGLPLSDIYKEGDHHPSERTKDGQYDPSIQQYEVSFKNNTVEGEYPSE SNYQVQEEPLSGAPNTWRRKKKKKTLDHRMNLIEHNLKDYSPMKDQLIHRKPLNSYISNF YDGTSDKSVQHRIGKIKLSTHRNDRRMIKDILTPQTVKVMKEYLLKRRKKYGNEKIQHLV DSSSDQMYREDSASLGNSWKGSIKVSHPLRFFPASFPLHTPMTANQGEAKR
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0111000 | 98 S | RSKSSTVDP | 0.995 | unsp | PKNH_0111000 | 98 S | RSKSSTVDP | 0.995 | unsp | PKNH_0111000 | 98 S | RSKSSTVDP | 0.995 | unsp | PKNH_0111000 | 95 S | PSVRSKSST | 0.991 | unsp | PKNH_0111000 | 97 S | VRSKSSTVD | 0.992 | unsp |