

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
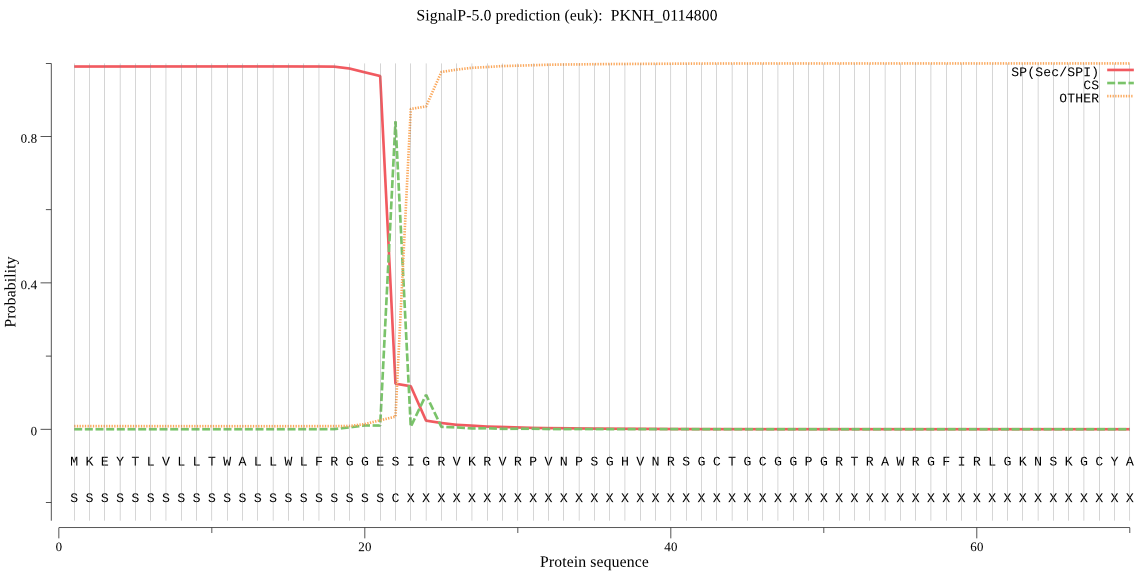
| PKNH_0114800 | SP | 0.007196 | 0.992648 | 0.000156 | CS pos: 22-23. GES-IG. Pr: 0.7835 |
MKEYTLVLLTWALLWLFRGGESIGRVKRVRPVNPSGHVNRSGCTGCGGPGRTRAWRGFIR LGKNSKGCYAKQRFPYRRQPERNHLTCATAGEGFSNTIHHDVKKNTQNYEEDGPPASFHR YIENIKTRKNVHPSIRITDFDKKFIRCKESYNRRYSHLKDSELFGNFRFVGRQKKGIMSP KYYLPKYVERPNYHRTGTPVPVPYEGERKMNSNTKGTESLHPYSNIKSEDDIETISSNCL FARELMDDVSHIICEGITTNDIDIYILNKCINNGYYPSPLNYHLFPKSSCISINEILCHG IPDNNVLYENDIVKVDISVFKDGFHADMCESFLVPKLSRNEKKKKKKFYDFIYLNDKLRT KYTKFIMKYNFDLTTNKVVKTGKQCSIRRIKYDPPNSKDRPNDQYNSFDDNTNCVMRSAH FDGNEITHEEDYYNDGNGATHMSRDNMQRDDLDELELFHRQYDEQVIYNKNQNSQTYDDI QRYIYNKTRGSPKDKNNRNHQNRFAFFDKGKLEADEFKMYMRKKNIDLIKTAHECTMEAI RVCKAGVPFSAIGDAIANHLDRINKRLHVHYAVVPHLCGHNIGKNFHEEPFIIHTRNNDD RQMCENLVFTIEPIISERPCDFIMWPDNWTMSNARYVFSAQFEHTIVVRKDGAEILTGKT DRSPKFVWECEET
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0114800 | 491 S | KTRGSPKDK | 0.998 | unsp | PKNH_0114800 | 491 S | KTRGSPKDK | 0.998 | unsp | PKNH_0114800 | 491 S | KTRGSPKDK | 0.998 | unsp | PKNH_0114800 | 156 S | NRRYSHLKD | 0.993 | unsp | PKNH_0114800 | 198 T | HRTGTPVPV | 0.994 | unsp |