

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0115100 | OTHER | 0.989841 | 0.003484 | 0.006675 |
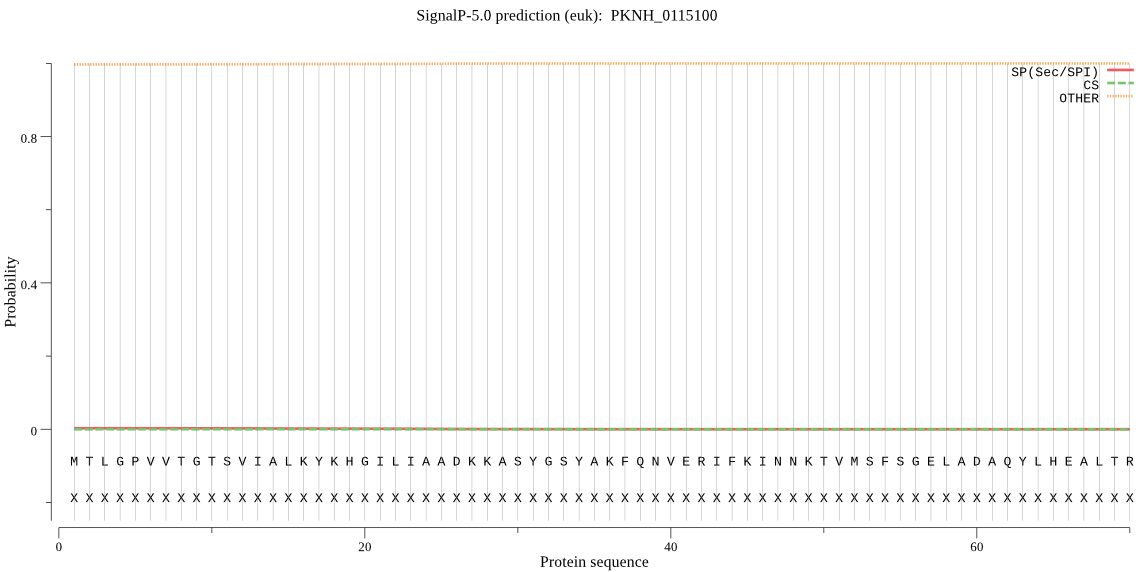
MTLGPVVTGTSVIALKYKHGILIAADKKASYGSYAKFQNVERIFKINNKTVMSFSGELAD AQYLHEALTRVNVNNVMDKKSKYDVHNTKYYHSYVGRLFYNRKNKIDPLFNTIIVAGVNS QDYDDNDKDVLLYSDKHTTDEYKIINKEDLYIGFVDMHGTQFSADYMTTGYARYFALTLL RDHYKDYMTEEEAKTLLNECLRVLYYRDATASNKIQIVKITSKGVEYEEPYFLSCDMNSR DYVYPSIMLPSTGCMW